- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Cell Signaling Journal
(Discontinued)
ISSN: 1876-3901 ― Volume 4, 2012
Evolution of the mob Gene Family
Xin Ye1, 2, §, Nikolas Nikolaidis1, 3, §, Masatoshi Nei1, 2, 3, Zhi-Chun Lai*, 1, 2, 3, 4
Abstract
Mob proteins from distantly related eukaryotic species share very high sequence similarity and they are characteristic of a conserved Mob domain with around 180 amino-acid residues in length. However, the evolutionary relationship of mob family genes has not been extensively investigated. Through a phylogenetic approach, we have conducted a comprehensive evolutionary analysis of the mob gene family. Here we show that over 270 mob family members from protists to animals can be organized in four distinct groups. This classification is strongly supported by the analysis of mob exon-intron structures. Moreover, the conservation and divergence patterns of different groups of Mob proteins have been elucidated. Structural information and the identification of fixed amino acid substitutions provide evidence about the putative significance of specific residues in the structural integrity and/or molecular functions of Mob proteins. Thus, this study reveals the evolutionary history of mob gene family and provides a basis for functional studies of Mob proteins
Article Information
Identifiers and Pagination:
Year: 2009Volume: 1
First Page: 1
Last Page: 11
Publisher Id: TOCELLSJ-1-1
DOI: 10.2174/1876390100901010001
Article History:
Received Date: 26/11/2008Revision Received Date: 17/12/2008
Acceptance Date: 19/12/2008
Electronic publication date: 14/1/2009
Collection year: 2009
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the 201 Life Sciences Building, The Pennsylvania State University, University Park, PA 16802, USA; Tel: (814) 863-0479; Fax: (814) 863-1357; E-mail: zcl1@psu.edu§ These authors contributed equally to this work.
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 26-11-2008 |
Original Manuscript | Evolution of the mob Gene Family | |
INTRODUCTION
The first member of the mob (Mps one binder) gene family, mob1, was identified in budding yeast as a critical regulator of mitosis [1]. In fission yeast, mob1 is involved in regulating processes such as cytokinesis [2]. These yeast Mob1 proteins bind to and stimulate the activities of Dbf2 and Sid2 protein kinases [3-5]. The other known Mob protein in yeast, Mob2, can similarly associate with and activate the kinase activities of Dbf2-related protein (Cbk1) and Sid2-related protein (Orb6) [6-8]. From these early studies, we began to understand that Mob proteins are critical intracellular signaling molecules and can function to regulate catalytic activity of certain protein kinases.
In human cells, Mob1-related proteins have been identified and shown to positively regulate Dbf2 homologues, NDR (nuclear Dbf2-related) protein kinases [9-1]. Similarly, the Drosophila Mob family proteins have been found to function as binding partners of fly Ndr family protein kinases Warts (Wts)/Lats (Large tumor suppressor) and Tricornered (Trc) [12, 13]. Moreover, the Drosophila mob1/mob as tumor suppressor (mats) (CG13852) was discovered as a growth inhibitor critical for cell proliferation and apoptotic control, and a human mats ortholog can functionally replace the fly mats gene to regulate tissue growth [12]. Studies on plant species also demonstrated the presence of Mob1-like genes and their putative function in cytokinesis [14-16]. Their cell cycle-regulated expression patterns and subcellular localizations indicate these Mob-like proteins function in cell proliferation and programmed cell death [16]. Therefore, Mob proteins are functionally important in both single cell and multi-cellular eukaryotes.
Mob proteins share high sequence similarity and they are characteristic of a conserved domain Mob1_phocein (pfam 03637) with around 180 amino acid residues in length (for simplicity, here we will call it the Mob domain). Mob family proteins are usually small and contain no other known structural motifs. The structural analyses of human, Xenopus laevis and yeast Mob1 proteins have revealed a central four-helix bundle stabilized by a zinc ion [17-19]. Moreover, they have identified an evolutionarily conserved acidic surface by which Mob proteins might interact with Ndr family protein kinases through electrostatic interactions.
Considering their high sequence similarity across distantly related species, we aimed to carry out a comprehensive evolutionary analysis to investigate the evolutionary relationship among all mob genes. Toward this goal, we have identified more than 270 mob family members from protists to animals, and conducted phylogenetic analysis. The study of the exon-intron structures elucidated the conservation and divergence patterns of different groups of mob genes. Structural information and the identification of fixed amino acid substitutions provided evidence about the putative significance of certain structural features or specific residues in the structural integrity and/or molecular functions of Mob proteins. Thus, this study offers not only the knowledge about the evolutionary history of mob gene family but also a basis for the prediction of biological functions of Mob proteins in eukaryotes.
MATERIALS AND METHODS
Extraction of Sequences
Mob protein sequences were obtained from the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nih.gov) and used as queries for BLASTp and tBLASTn searches against NCBI databases (http://www.ncbi.nih.gov) and Ensembl databases (http://www.ensembl.org/index.html). Some Mob proteins were identified by searching genome sequences from organism-specific websites (Dictybase - http://dictygenome.org, the Giardia lamblia Genome Database - www.mbl.edu/Giardia, and Ciona intestinalis Genome - http://genome.jgi-psf.org/Ciona). Only sequences that have more than 50 percent coverage of query lengths were kept. Truncated sequences and redundant sequences were eliminated. The final 73 sequences used for this study are summarized in Supplementary Table S1 with information on species, gene names and accession numbers.
Sequence Alignments
Initially, full-length protein sequences were aligned using Multiple Alignment Mode in ClustalX1.83 with default parameter setting. We used MEGA3.1 to generate a preliminary neighbor-joining (NJ) tree for observing a classification pattern of Mob family. Both N-terminus and C-terminus were shown to have poor sequence conservation compared to the Mob domain region. This is the only domain found in Mob proteins and it comprises about 80 percent of the full protein length. We used Mob domain sequences for further phylogenetic analysis. We also did individual alignment for each subgroup and profile alignments (ClustalX) for other analyses, such as identification of conserved substitutions and conservation mapping.
Phylogenetic Analysis
Multiple alignment results of Mob domain sequences were subject to neighbor-joining and maximum parsimony phylogeny reconstruction using MEGA3.1 and PHYLIP. NJ tree was rooted with a Mob from Diplomonadida (Giardia lambia) and constructed using p-distances with complete elimination of alignment gaps. One hundred forty-four amino acids of the Mob domain were used. The reliability of the resulting NJ and MP trees was tested by 1000 bootstrap resamplings.
Exon-Intron Structures
The exon-intron structures of mob genes from representative organisms were determined by mRNA-to-genomic alignment using Spidey program (http://www.ncbi.nlm.nih.gov/IEB/Research/Ostell/Spidey/spideyweb.cgi) or directly obtained from Ensembl web pages.
Expression Analysis of Human MOB Genes
The expression data of H1 histone member 0 (H1_0) and all human MOB genes were obtained from EST profiles of UniGene in NCBI. Updated EST data was collected in August of 2008 (Table S2).
RESULTS
mob Genes are Present Widely in Eukaryotes
Since the discovery of the first mob family member in yeast [1], more than 270 mob genes have been found in a variety of eukaryotes from the primitive protists to plants and animals. Two mob genes, mob1 and mob2, exist in single cell eukaryotes such as budding yeast S. cerevisiae. Neurospora has one more mob gene in addition to the mob1-like and mob2-like genes. Protists such as Giardia have at least three mob genes, which is comparable to the mob gene number in invertebrates D. melanogaster (four) and C. elegans (four). Generally, vertebrates have more mob family members. For example, there are seven mob genes in the human, mouse, rat, zebrafish and fugu genomes, eight in frog and five in chicken genomes. Because mob genes are only found in eukaryotes, the mob gene family appears to be an innovation of eukaryotic organisms.
Evolutionary Relationships of mob Family Genes
Phylogenetic analysis identified four major groups of mob genes (Fig. 1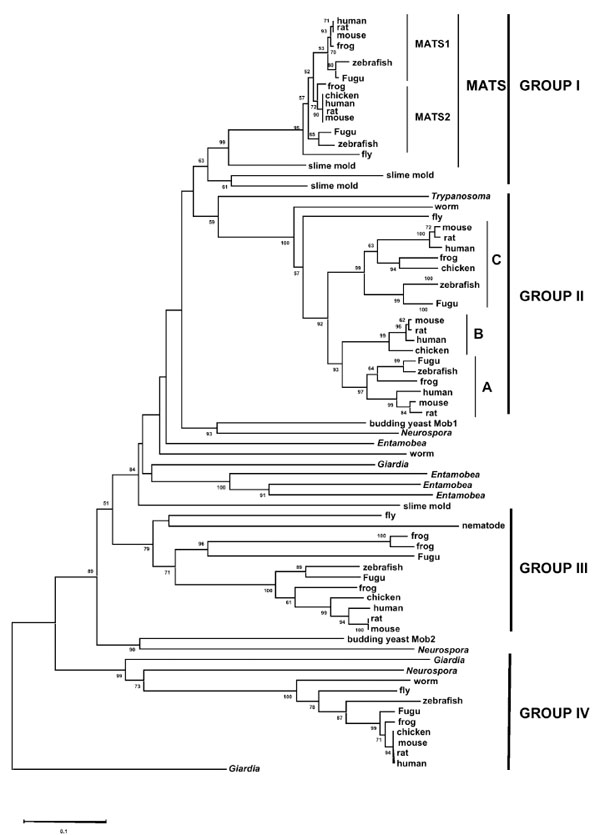 ). Protists such as Giardia and Entamoeba have mob genes in almost all groups, indicating that gene duplications giving rise to major mob family members occurred very early in evolution. Although each group has only one mob gene from invertebrates, Group I and Group II have at least two mob genes from most vertebrates. It suggests that additional gene duplications have resulted in the different mob genes repertoire of vertebrates.
). Protists such as Giardia and Entamoeba have mob genes in almost all groups, indicating that gene duplications giving rise to major mob family members occurred very early in evolution. Although each group has only one mob gene from invertebrates, Group I and Group II have at least two mob genes from most vertebrates. It suggests that additional gene duplications have resulted in the different mob genes repertoire of vertebrates.
Group I, also called Mats group [12], can be further divided into two subgroups, Mats1 (also named Mobkl1b for Mps one binder kinase activator-like 1b) and Mats2 (also named Mobkl1a for Mps one binder kinase activator-like 1a) (Fig. 1 ). Both of these two subgroups contain vertebrate mob genes from fish to mammals except for the chicken mats1, which appears to be lost in a lineage-specific manner. Fruit flies and mosquito, however, only have one mob gene in this group, which is supportive of additional gene duplication event occurring in vertebrates after their divergence from invertebrates. Interestingly, no mats gene is found in nematodes C. elegans and C. briggsae. This could be due to a lineage-specific gene loss in nematodes.
). Both of these two subgroups contain vertebrate mob genes from fish to mammals except for the chicken mats1, which appears to be lost in a lineage-specific manner. Fruit flies and mosquito, however, only have one mob gene in this group, which is supportive of additional gene duplication event occurring in vertebrates after their divergence from invertebrates. Interestingly, no mats gene is found in nematodes C. elegans and C. briggsae. This could be due to a lineage-specific gene loss in nematodes.
Group II contains three clusters of vertebrate mob genes but only one homolog from fly and worm (Fig. 1 ). Two additional mob genes in vertebrates should have resulted from two rounds of gene duplications in the common ancestor of vertebrate lineages. In these three clusters of vertebrate mob genes, however, one cluster does not have members from zebrafish and Fugu. This implies a putative gene loss from the fish lineages.
). Two additional mob genes in vertebrates should have resulted from two rounds of gene duplications in the common ancestor of vertebrate lineages. In these three clusters of vertebrate mob genes, however, one cluster does not have members from zebrafish and Fugu. This implies a putative gene loss from the fish lineages.
Group III, different from Group I and Group II, has only one mob gene from both invertebrates and vertebrates (Fig. 1 ). Interestingly, two frog and one fugu mob genes form an additional cluster with relatively long-branch lengths. Since the probability of gene losses simultaneously occurring in zebrafish, chicken and mammals is low, it is likely that fugu and frog have had some lineage-specific gene duplications. These additional frog and fugu genes might have been subject to pseudogenization, which is supported by highly diverged protein sequences as inferred from the long-branch lengths. Based on the observation of lineage-specific gene losses and/or gene duplications and the clustering of orthologs instead of paralogs, all these three groups of mob genes seem to have experienced the birth-and death model of evolution [20]. Finally, Group IV, which contains single mob gene from each species, is considered to be the closest one to outgroup mob genes (Fig. 1
). Interestingly, two frog and one fugu mob genes form an additional cluster with relatively long-branch lengths. Since the probability of gene losses simultaneously occurring in zebrafish, chicken and mammals is low, it is likely that fugu and frog have had some lineage-specific gene duplications. These additional frog and fugu genes might have been subject to pseudogenization, which is supported by highly diverged protein sequences as inferred from the long-branch lengths. Based on the observation of lineage-specific gene losses and/or gene duplications and the clustering of orthologs instead of paralogs, all these three groups of mob genes seem to have experienced the birth-and death model of evolution [20]. Finally, Group IV, which contains single mob gene from each species, is considered to be the closest one to outgroup mob genes (Fig. 1 ). All four groups of mob genes diverged following the species tree, which is indicative of divergent evolution rather than concerted evolution.
). All four groups of mob genes diverged following the species tree, which is indicative of divergent evolution rather than concerted evolution.
Exon-Intron Structures of mob Genes
To further reveal evolutionary relationships among mob genes, we analyzed and compared exon-intron structures within the coding region of mob genes from different groups
(Fig. 2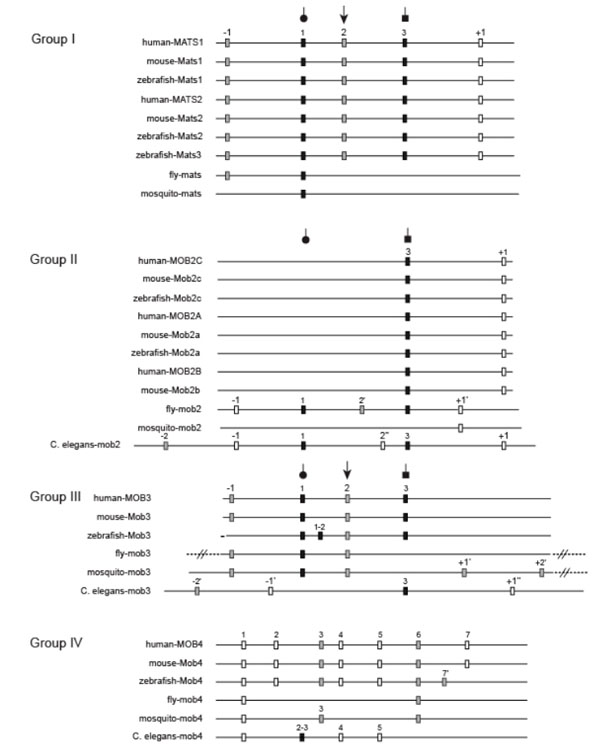 ). For vertebrate mob genes, the numbers, positions and phases of introns are generally conserved within the same group with two exceptions. One is Group III zebrafish mob, which has an additional intron1-2 and is lack of the intron-1, and the other one is Group IV zebrafish mob, whose last intron has a different position and phase from other vertebrate orthologous mob genes. In the central region of mob sequences, which corresponds to the Mob domain, Group I, II and III mob genes have similar exon-intron structures whereas Group IV mob genes are quite different and share their own unique splicing pattern. The inter-group comparison among Group I, II and III shows a higher conservation level of intron properties in the Mob domains than either amino-terminal or carboxyl-terminal regions.
). For vertebrate mob genes, the numbers, positions and phases of introns are generally conserved within the same group with two exceptions. One is Group III zebrafish mob, which has an additional intron1-2 and is lack of the intron-1, and the other one is Group IV zebrafish mob, whose last intron has a different position and phase from other vertebrate orthologous mob genes. In the central region of mob sequences, which corresponds to the Mob domain, Group I, II and III mob genes have similar exon-intron structures whereas Group IV mob genes are quite different and share their own unique splicing pattern. The inter-group comparison among Group I, II and III shows a higher conservation level of intron properties in the Mob domains than either amino-terminal or carboxyl-terminal regions.
Group I:
Vertebrate mats genes have the same splicing pattern which produces 6 exons, while Drosophila and mosquito only have three and two exons respectively. Intron2 (phase 2) and intron3 (phase 1) have been lost in invertebrate lineages. Vertebrate mats genes have an additional intron+1 (phase 0) inserted in the C terminal region. Since this intron is not present in any other mob genes, it might be due to either a gain-of-intron in vertebrate mats, or, that the last intron actually arose in the common ancestor of vertebrate and invertebrate mats genes and then got lost in invertebrates. In addition, intron1, 2 and 3 must have been existent before the gene duplications that produced Group I and Group III mob genes since they are all present in most members of these two groups.
Group II:
Different from Group I mob genes, invertebrate members of this group generally have more introns (five for D. melanogaster and six for C. elegans) than vertebrates (two) except for mosquito mob2 that only has one intron. The two introns from vertebrate mob2 genes are the conserved intron 3 and an intron+1 located further downstream. Both of these two introns are shared by C. elegans. Immediately downstream to this intron+1, there exists Lys/Arg as the first amino acid of the flanking exon. The fly and mosquito mob2 genes, which are lack of this conserved intron+1 in the corresponding positions, have Gln instead of Lys/Arg in the corresponding site. Coincidently, in mats genes presented in Fig. (2 ), Gln is shared by mats genes, from which this conserved intron+1 is absent. This might be explained by the gain-of-intron in Group II vertebrate and C. elegansmob genes which was triggered by the amino acid substitution from Gln to Lys / Arg. Alternatively, it could have happened via intron loss in Group I mats and Group II insect mob respectively, following the substitution from Lys/Arg to Gln. In addition to these two introns shared with vertebrates, C. eleganmob2 has four more introns, two of which are conserved in fly mob2 (intron-1 and intron1). Surprisingly, mosquito mob2 only has one intron (intron+1’). This intron can be found in fly too and it might have been produced exclusively in insects. All other upstream introns have been lost in this species.
), Gln is shared by mats genes, from which this conserved intron+1 is absent. This might be explained by the gain-of-intron in Group II vertebrate and C. elegansmob genes which was triggered by the amino acid substitution from Gln to Lys / Arg. Alternatively, it could have happened via intron loss in Group I mats and Group II insect mob respectively, following the substitution from Lys/Arg to Gln. In addition to these two introns shared with vertebrates, C. eleganmob2 has four more introns, two of which are conserved in fly mob2 (intron-1 and intron1). Surprisingly, mosquito mob2 only has one intron (intron+1’). This intron can be found in fly too and it might have been produced exclusively in insects. All other upstream introns have been lost in this species.
Group III:
Group III mob genes remain most of the three conserved introns (intron1, 2, and 3). Intron-1 in this group is in the similar but not the same position as intron-1 in Group I. Zebrafish gained an additional intron1-2 compared to other vertebrates. Fly and mosquito both lost intron3, which is, however, the only intron from vertebrates that is present in C. elegans as well. Except for this intron, C. elegansmob3 has a very different exon-intron structure compared to other group III members.
Group IV:
mob genes in this group are unique in their splicing patterns. They don’t have any of the three conserved introns (intron1, 2, and 3) from other three groups. Vertebrates of this group share mostly the same intron properties except that zebrafish has a different intron7. Intron1, 3, 4, 5 and 6 are shared by either some or all of the three invertebrate species. Overall, vertebrates have more introns than invertebrates in this group.
Based on the exon-intron structures of four groups of mob genes, we can infer the mob family evolution pattern in terms of gene duplications. The first gene duplication produced two mob genes. One evolved all the way to the contemporary Group IV mob. The other one duplicated again, giving rise to Group III and the common ancestor of Group I and Group II. Later on, another round of gene duplication occurred which gave birth to Group I and Group II mob genes. These three gene duplication events all should have occurred before the divergence of vertebrates and invertebrates since fly, mosquito and nematode homologs are all present in these four groups except that nematode mats is suspected to be lost during evolution. Combined with the phylogenetic tree, which shows that Giardia mob genes are present in almost all groups (Fig. 1 ), we can even trace the original three gene duplication events to the very early stage of eukaryote evolution. mob genes from fungi are present in Group IV, Group III and somewhere between Group I and II. This pattern also supports the order in which these four Mob groups have been established.
), we can even trace the original three gene duplication events to the very early stage of eukaryote evolution. mob genes from fungi are present in Group IV, Group III and somewhere between Group I and II. This pattern also supports the order in which these four Mob groups have been established.
Both the sequence alignment and exon-intron structure showed that the N-terminal regions of Mob proteins are quite variable for the inter-group comparison but highly conserved within individual groups. Therefore, these regions have diverged more rapidly than other regions after separation of different groups. Since these regions appear to be unique to specific group, it is indicative of strong positive selection and suggests an important role of this region in group-specific functions.
Structural Conservation of Mob Proteins
Stavridi et al. [17] resolved the X-ray crystal structure of human Group I MOB1A/MATS1 protein and this structure has been used as a template in our analysis. Human MOB1A protein folds into a four-helix bundle core structure, which is stabilized by a Cys2His2 zinc finger holding a zinc atom in the middle. The removal of zinc using EDTA was shown to lead to the aggregation of the protein and decreases of its thermal stability [18]. On the surface of hMOB1A protein, one side is flat and rich in negative electrostatic potential. Several studies have reported that Mob proteins can interact with NDR family kinases and stimulate their kinase activity [21]. The structural analysis of NDR family kinases has identified at least two conserved basic regions, and this acidic surface of hMOB1A protein appears to be functionally important by binding with the conserved basic regions of NDR family kinases [17].
To examine how structures of four groups of Mob proteins are conserved compared to hMOB1, the program ConSurf was used to map amino acids of each group of Mob proteins and colored amino acid residues according to their conservation levels by using the structure of hMOB1 as template (Fig. 3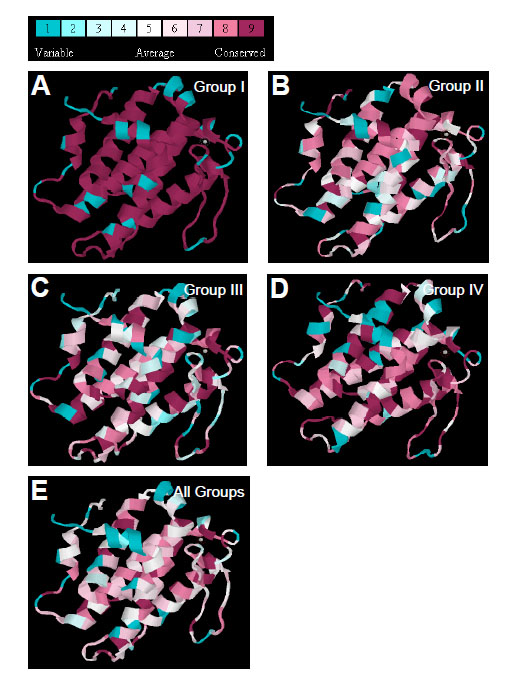 ). Overall, Mob proteins are highly conserved in their amino acids sequences, among which Group I members have the highest conservation level as expected (Fig. 3A
). Overall, Mob proteins are highly conserved in their amino acids sequences, among which Group I members have the highest conservation level as expected (Fig. 3A ). Group II, although not as conserved as Group I, preserve high sequence similarity in the C-terminal of H2, C-terminal of H5, and four residues of the Cys2His2 structure (Fig. 3B
). Group II, although not as conserved as Group I, preserve high sequence similarity in the C-terminal of H2, C-terminal of H5, and four residues of the Cys2His2 structure (Fig. 3B ). Group III and Group IV Mobs are both conserved in their zinc finger regions and the flanking structural elements, including N terminal of H4, C-terminal of H5 and some residues from H2 (Fig. 3C
). Group III and Group IV Mobs are both conserved in their zinc finger regions and the flanking structural elements, including N terminal of H4, C-terminal of H5 and some residues from H2 (Fig. 3C , D
, D ). Thus, comparison of all four groups of Mob proteins uncovered the highly conserved zinc finger structure and its closely positioned motifs, and also some residues that are critical for stabilizing loop structures.
). Thus, comparison of all four groups of Mob proteins uncovered the highly conserved zinc finger structure and its closely positioned motifs, and also some residues that are critical for stabilizing loop structures.
To identify residues that are highly conserved among different groups, human, mouse, zebrafish and fly Mob proteins were used to make alignments (Fig. 4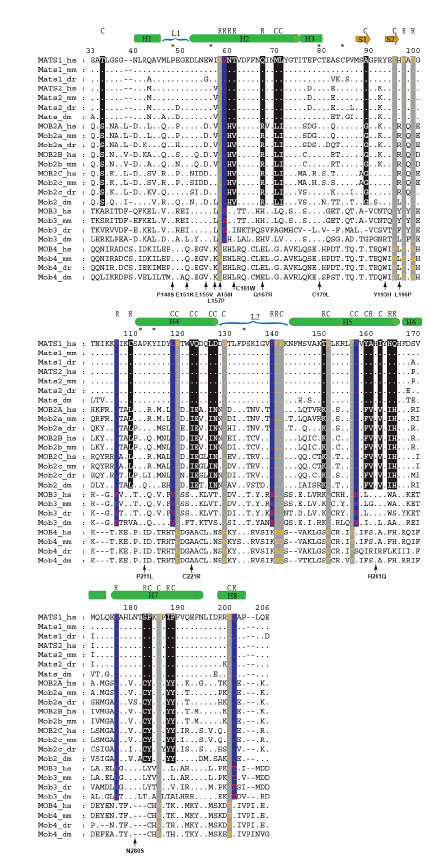 ). A total of seven residues are conserved in all four groups of vertebrate and fly Mobs. Pro48 from L1 and Pro133 from L2 may play an important role in stabilizing the loop structure and further maintaining the folding of core four-helix bundle. Moreover, the N-terminals of H2 and H4 helices have the conserved Trp56, Ala111 and Tyr114, all of which are hydrophobic residues. Trp56 is shown to have buried side chains [17]. Ala111 and Tyr114 are spatially close to zinc finger motif and facing inward to the core of helix bundle, which may contribute to the protein stability by hydrophobic interactions. Two remaining conserved residues are Cys79 and Cys84. These two cysteins, together with His161 and His166, coordinate a zinc atom. His161 and His 166 are also highly conserved with the exception of zebrafish Mob4, in which these two histidines are both replaced by isoleucines. The ubiquitous presence of this Cys2His2 zinc finger indicates that it is critical structural feature for Mob family proteins.
). A total of seven residues are conserved in all four groups of vertebrate and fly Mobs. Pro48 from L1 and Pro133 from L2 may play an important role in stabilizing the loop structure and further maintaining the folding of core four-helix bundle. Moreover, the N-terminals of H2 and H4 helices have the conserved Trp56, Ala111 and Tyr114, all of which are hydrophobic residues. Trp56 is shown to have buried side chains [17]. Ala111 and Tyr114 are spatially close to zinc finger motif and facing inward to the core of helix bundle, which may contribute to the protein stability by hydrophobic interactions. Two remaining conserved residues are Cys79 and Cys84. These two cysteins, together with His161 and His166, coordinate a zinc atom. His161 and His 166 are also highly conserved with the exception of zebrafish Mob4, in which these two histidines are both replaced by isoleucines. The ubiquitous presence of this Cys2His2 zinc finger indicates that it is critical structural feature for Mob family proteins.
Divergence of Mob Proteins
Structural diversification would allow functional divergence of Mob proteins. Group I and group II have 22 fixed substitutions, most of which are located in helix regions (19 out of 22) (Fig. 4 ). Radical substitutions in helices are distributed in H2, H5 and H7 with the numbers of 3, 4 and 2 for each. Based on the phylogeny relationships of mob genes, which indicate that Groups I and II were produced following Group IV and Group III, His60 was suspected to have been replaced by Asn in the common ancestral sequence of Mob1, Mob2 and Mob3 and then after the duplication that generated Mob1 and Mob2, the relaxed functional constraints on one of the two duplicates (Mob2 in this case) made the backward mutation (N->H) possible. The neighboring residue Thr61 might have been subject to the similar evolution course, where Mob1 and most Mob3 share Thr while Mob2 and Mob4 contain conservative residues, Val and Leu, respectively. The third radical substitution (Q67R), however, is different from the previous two in that Mob2 and Mob3 are now sharing the conservative positively charged amino acids, Arg and His, but Mob1 and Mob4 have Gln and Glu instead. The substitution of T150K from H5 happened in the same way as Q67R, where Mob1/Mob4 has Thr/Ser but Mob2 and Mob3 share Lys. H164I and Q165H are next to each other and these two sites function to position H5 and H6 together in a perpendicular direction. The interaction between these two residues might be required to maintain this particular angle. Based on the fact that these two sites always carry the opposite electrostatic charges and even if radical substitutions occurred, they occurred in a compensatory way (I->H, H->Q), the electrostatic interaction between them is very likely to be critical for the structural integrity. The A160V substitution, although it is radical, might not have dramatic effect since both Ala and Val are neutral and only differ in their volume. The last two radical changes in helices are located in the middle region of H7 and both of them come together with the flanking conservative F-to-Y substitution. The only radical substitution (C109L) in non-helix region is located N terminally to H4. Since Cys is generally conserved in Groups I, III and IV, the presence of Leu in Group II is supposed to have arisen in a group-specific way and might be critical for Group II specificity.
). Radical substitutions in helices are distributed in H2, H5 and H7 with the numbers of 3, 4 and 2 for each. Based on the phylogeny relationships of mob genes, which indicate that Groups I and II were produced following Group IV and Group III, His60 was suspected to have been replaced by Asn in the common ancestral sequence of Mob1, Mob2 and Mob3 and then after the duplication that generated Mob1 and Mob2, the relaxed functional constraints on one of the two duplicates (Mob2 in this case) made the backward mutation (N->H) possible. The neighboring residue Thr61 might have been subject to the similar evolution course, where Mob1 and most Mob3 share Thr while Mob2 and Mob4 contain conservative residues, Val and Leu, respectively. The third radical substitution (Q67R), however, is different from the previous two in that Mob2 and Mob3 are now sharing the conservative positively charged amino acids, Arg and His, but Mob1 and Mob4 have Gln and Glu instead. The substitution of T150K from H5 happened in the same way as Q67R, where Mob1/Mob4 has Thr/Ser but Mob2 and Mob3 share Lys. H164I and Q165H are next to each other and these two sites function to position H5 and H6 together in a perpendicular direction. The interaction between these two residues might be required to maintain this particular angle. Based on the fact that these two sites always carry the opposite electrostatic charges and even if radical substitutions occurred, they occurred in a compensatory way (I->H, H->Q), the electrostatic interaction between them is very likely to be critical for the structural integrity. The A160V substitution, although it is radical, might not have dramatic effect since both Ala and Val are neutral and only differ in their volume. The last two radical changes in helices are located in the middle region of H7 and both of them come together with the flanking conservative F-to-Y substitution. The only radical substitution (C109L) in non-helix region is located N terminally to H4. Since Cys is generally conserved in Groups I, III and IV, the presence of Leu in Group II is supposed to have arisen in a group-specific way and might be critical for Group II specificity.
Between Groups I/II and Group III, there are totally seven fixed amino acid substitutions in the Mob domain and five of them are radical changes (Fig. 4 ). Among these five changes, two are located in the non-helix regions, which might be critical for group-specific functions. P106K, E176H and L201T could be due to substitutions specifically occurring in Group III since the rest three groups share the same amino acids. P139E, however, might be the result of two independent substitutions in Groups I/II (P) and Group III (E) from their ancestral gene, or Glu is the ancestral residue and Pro has derived from it. The last radical change is V59S from H2. This region has four continuous radical substitutions (site 58-61), which cover all three categories. It may indicate the importance of this region in the functional differentiation of different groups of Mob proteins. For two conservative substitutions (L118V, R157H), both occur in helix regions and may have less effect.
). Among these five changes, two are located in the non-helix regions, which might be critical for group-specific functions. P106K, E176H and L201T could be due to substitutions specifically occurring in Group III since the rest three groups share the same amino acids. P139E, however, might be the result of two independent substitutions in Groups I/II (P) and Group III (E) from their ancestral gene, or Glu is the ancestral residue and Pro has derived from it. The last radical change is V59S from H2. This region has four continuous radical substitutions (site 58-61), which cover all three categories. It may indicate the importance of this region in the functional differentiation of different groups of Mob proteins. For two conservative substitutions (L118V, R157H), both occur in helix regions and may have less effect.
The comparison between Groups I/II/III and Group IV identifies four radical substitutions and eight conservative ones (Fig. 4 ). The radical changes are mainly limited to non-helix regions (3 out of 4). The only radical substitution on helix is A58Y from H2. This residue Ala was replaced by Ile in one of the yeast conditional mob1 mutants (mob1-95) [1]. Thus, this Ala might be critical for some specific function shared by Groups I, II and III but not by Group IV. The rest three radical changes are located C terminally to the two beta strands (W97C, D99A) and N terminally to H5 (F140E). Phe140 was thought to be involved in structural interaction, which together with Phe132 and Phe144 form hydrophobic interactions with each other and also with conserved Ile151 from H5 [17].
). The radical changes are mainly limited to non-helix regions (3 out of 4). The only radical substitution on helix is A58Y from H2. This residue Ala was replaced by Ile in one of the yeast conditional mob1 mutants (mob1-95) [1]. Thus, this Ala might be critical for some specific function shared by Groups I, II and III but not by Group IV. The rest three radical changes are located C terminally to the two beta strands (W97C, D99A) and N terminally to H5 (F140E). Phe140 was thought to be involved in structural interaction, which together with Phe132 and Phe144 form hydrophobic interactions with each other and also with conserved Ile151 from H5 [17].
A number of mutations in the yeast mob1 genes have been molecularly characterized, most of which are located in H2 helix, especially in its N-terminal, and also L1 loop (Stavridi et al. 2003). The other ones are scattered on H4, C-terminal of H5 and N-terminal of H7. Also some are residing in the non-helix region that connects H2 and H3. The mutations of E151K (mob1-77), Q167R (mob1-55) and Y193H (mob1-55) can cause late mitotic arrest and cytokinesis defects although these residues are only conserved in Group I and/or Group II Mobs but not in Group III ones. It suggests that Mob1 might have distinct function from Group III Mobs or have acquired new mechanism that makes these residues essential for Mob1 but not for Mob3. The other mob1 alleles mostly have mutations occurring in conservative residues shared by Mob1 and Mob3, indicating their significant role in the common features of Group I and Group III Mobs.
Human mob Genes are Generally Expressed in Most Tissues and During Development
At present time, very little is known about the functions of most mob family genes. Analysis of mob gene expression would help understand how mob genes might be functionally required in different tissues during development. For this reason, we have focused on human MOB genes to elucidate their expression profiles since abundant Expression Sequence Tag (EST) data is available from UniGene database in NCBI. Overall expression of hMATS1 during development is the highest among all hMOB genes and is almost comparable to that of control gene, H1 histone member 0 (H1_0) (Fig. 5A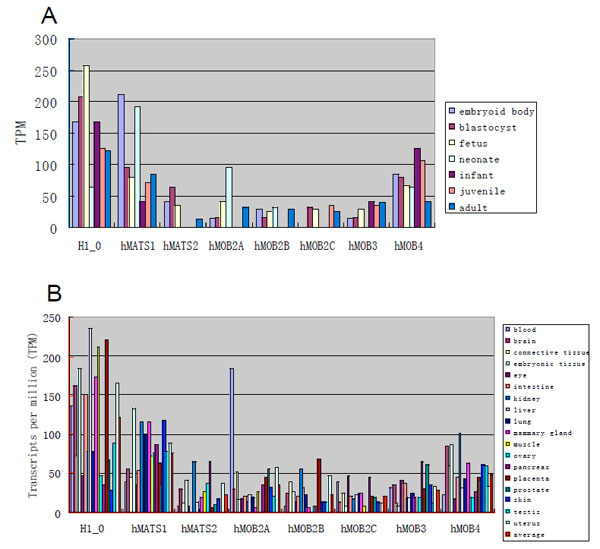 ). Human MOB genes are generally expressed throughout development except that some members show zero EST in neonate, infant or juvenile stage (Fig. 5A
). Human MOB genes are generally expressed throughout development except that some members show zero EST in neonate, infant or juvenile stage (Fig. 5A ). Considering that there are only 5x104 or less total ESTs from each of these three stages, the failure to detect MOB-related ESTs might not accurately reflect expression of the MOB genes.
). Considering that there are only 5x104 or less total ESTs from each of these three stages, the failure to detect MOB-related ESTs might not accurately reflect expression of the MOB genes.
To analyze human MOB expression in different tissues, only the tissues that have more than 105 of total ESTs were used. Generally, all hMOB genes are expressed in most tissues at levels lower than H1_0 gene (Fig. 5B ). Among the seven hMOB genes, hMATS1 has the highest average expression. hMOB2A has outstanding expression in blood, which is five times higher than its average level. All other hMOB genes, however, are mostly expressed within two times of their average levels. Expression of hMATS2, hMOB2B and hMOB4 in kidney and expression of hMATS2, hMOB2C and hMOB3 in pancreas are more than twice higher than their average levels. Moreover, placenta, eye and prostate show more than twice higher expression levels for hMOB2B, hMOB2C and hMOB3, respectively. Interestingly, some tissues show no expression for some of the hMOB genes, such as hMATS2 in intestine and testis, hMOB2B and hMOB3 in muscle, as well as hMOB2A and hMOB2C in ovary. Thus, while hMOB genes are generally expressed in most tissues, some of them appear to be preferentially expressed in certain tissues.
). Among the seven hMOB genes, hMATS1 has the highest average expression. hMOB2A has outstanding expression in blood, which is five times higher than its average level. All other hMOB genes, however, are mostly expressed within two times of their average levels. Expression of hMATS2, hMOB2B and hMOB4 in kidney and expression of hMATS2, hMOB2C and hMOB3 in pancreas are more than twice higher than their average levels. Moreover, placenta, eye and prostate show more than twice higher expression levels for hMOB2B, hMOB2C and hMOB3, respectively. Interestingly, some tissues show no expression for some of the hMOB genes, such as hMATS2 in intestine and testis, hMOB2B and hMOB3 in muscle, as well as hMOB2A and hMOB2C in ovary. Thus, while hMOB genes are generally expressed in most tissues, some of them appear to be preferentially expressed in certain tissues.
DISCUSSION
The mob Gene Family Has Four Distinct Groups
Up to date, there are more than 270 mob genes identified from eukaryotes. Through a molecular evolutionary approach, we have elucidated evolutionary relationships among these mob genes. It is clear that there are four distinct groups of mob genes (Groups I-IV), which should have evolved before the divergence of vertebrate and invertebrate animals. A similar conclusion was reached in a previous evolutionary analysis done with 192 mob genes [22]. Like other gene families, gene duplication provided a mechanism for generating new family members. Because protists have mob genes in almost all groups, the first three gene duplication events should have occurred at a very early stage of eukaryotic evolution. As mob gene is only found in eukaryotes, the mob genes should have arisen after the divergence of prokaryotes and eukaryotes and the mob gene family appears to be an innovation of eukaryotic organisms.
Conserved Features of Mob Proteins
Mob proteins appear to share the following three major structural features. The first feature is that Mob has an atypical Cyc2-His2 motif responsible for zinc binding [17]. This is a general property of all Mob proteins probably with an exception of zebrafish Mob4 (Fig. 4 ). The second feature is that there is a flat surface on one side of Mob protein rich in acidic residues. This structure is presumably critical for Mob1 to interact with its partner such as NDR family protein kinases. The third feature is that generally Mob proteins are small in size. In addition to the Mob domain, there are no other obvious domains in Mob proteins. While it is common for a protein domain to be linked with other domains in a protein to increase structural and functional complexity, it is not clear why Mob proteins are restricted to increase their size and not allowed to combine with other protein domains. As Mob protein can associate with other proteins such as Ndr kinase, it is possible that there is a space constriction for Mob to fit into a protein complex, which prevents Mob from altering its overall size.
). The second feature is that there is a flat surface on one side of Mob protein rich in acidic residues. This structure is presumably critical for Mob1 to interact with its partner such as NDR family protein kinases. The third feature is that generally Mob proteins are small in size. In addition to the Mob domain, there are no other obvious domains in Mob proteins. While it is common for a protein domain to be linked with other domains in a protein to increase structural and functional complexity, it is not clear why Mob proteins are restricted to increase their size and not allowed to combine with other protein domains. As Mob protein can associate with other proteins such as Ndr kinase, it is possible that there is a space constriction for Mob to fit into a protein complex, which prevents Mob from altering its overall size.
Since mob genes are so highly conserved, they are expected to play important roles in establishing and maintaining key features of eukaryotes during evolution. In one case, genetic analysis has shown that Drosophila mats gene plays an essential role in cell proliferation and apoptotic control during tissue growth and a human MATS gene can functionally replace fly mats [12]. At the molecular level, Mats functions as a binding partner and coactivator of Wts/Lats protein kinase [12]. Since the Drosophila Mats protein has been shown to function as a growth inhibitor to control tissue growth during development, we speculate that loss of MATS function might promote tumorigenesis of human cancers. Furthermore, activation of Ndr family kinases by Mob has been demonstrated in wide variety of species from yeast to humans [reviewed in 21]. Importantly, yeast Mob2 also binds NDR family protein kinases, Cbk1 and Orb6, to control polarized cell growth [reviewed in 21]. Therefore, this conserved function for Mob as kinase coactivator can be at least traced back to the time when the first mob gene duplication occurred. Investigation of whether Group IV Mob can function as a kinase activator would help investigate the possibility of this molecular feature being innovated at the very beginning of mob gene evolution.
Functional Diversification of Mob Proteins
Structural alterations make it possible for functional diversification. To characterize structural differences among four groups of Mob proteins, both radical and conservative substitutions between different groups have been examined. As summarized in Table 2, both types of substitutions can occur in helix as well as non-helix regions (Fig. 4 ). Moreover, substitutions occurred throughout Mob protein from the amino to carboxyl termini. In one example, Thr74 is a conserved residue among Group I/II/III Mob proteins with the exception of zebrafish Mob3, but it is replaced by a Lys in Group IV (Fig. 4
). Moreover, substitutions occurred throughout Mob protein from the amino to carboxyl termini. In one example, Thr74 is a conserved residue among Group I/II/III Mob proteins with the exception of zebrafish Mob3, but it is replaced by a Lys in Group IV (Fig. 4 ). This residue was shown to be phosphorylated by MST2 protein kinase and important for MOB1 to activate NDR1 protein kinase [23]. Through this mechanism, MST2 protein kinase functions as an upstream regulator of Group I/II/III but not Group IV Mob proteins. As we begin to understand the importance of phosphorylation for Mob regulation [23-25], conserved sequence changes would allow Mob proteins of various groups to be differently regulated by phosphorylation and other protein modification mechanisms.
). This residue was shown to be phosphorylated by MST2 protein kinase and important for MOB1 to activate NDR1 protein kinase [23]. Through this mechanism, MST2 protein kinase functions as an upstream regulator of Group I/II/III but not Group IV Mob proteins. As we begin to understand the importance of phosphorylation for Mob regulation [23-25], conserved sequence changes would allow Mob proteins of various groups to be differently regulated by phosphorylation and other protein modification mechanisms.
ACKNOWLEDGEMENTS
This work was partly supported by an National Institutes of Health grant to M.N. and an National Science Foundation grant to Z.-C.L.







