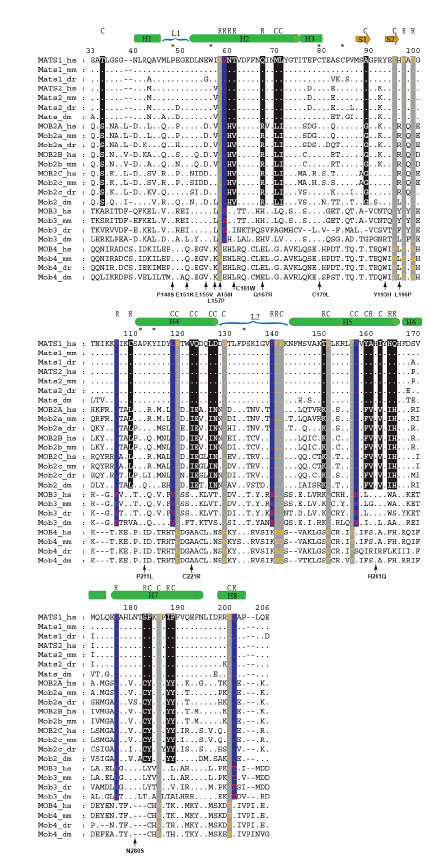
Fig. (4) Sequence alignment of vertebrate and fly mob proteins. Human, mouse, zebrafish and fly Mob proteins were used to make profile
alignments and the sequences cover residues 33 to 206 of human MOB1, in which the main motifs of the crystal structure of
hMOB1A/hMATS1 protein were displayed (Stavridi et al. 2003). The sequences analyzed here cover the entire Mob domain. The protein
names follow the nomenclature described in Table 1. We use dots for conserved residues and dashes for alignment gaps. The numbers on top
of the alignment blocks reflect the actual amino acid positions of hMATS1. Asterisks are used to locate conserved residues for all aligned
sequences. The secondary structures of hMATS1 are shown as: green bar-helix, brown arrow-beta sheet, blue parenthesis-loop. The fixed
amino acid substitutions are highlighted by white fonts in black background (Group I vs Group II), red fonts in blue background (Groups I/II
vs Group III), and orange fonts in gray background (Groups I /II/III vs Group IV). The properties of these substitutions are indicated on top
of the corresponding sites, R for radical substitution and C for conservative substitution. The amino acid substitutions encoded by S. cerevisiae
mob1 mutant alleles (Luca and Winey 1998, Stavridi et al. 2003) are positioned to the corresponding sites at the bottom of alignment
blocks. The numbers used for the mutants’ names are the actual residue positions of yeast Mob1 protein. The letters flanking the left and
right sides of these numbers indicate the amino acids before and after mutations, respectively.