- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Drug Metabolism Journal
(Discontinued)
ISSN: 1874-0731 ― Volume 5, 2011
In Vitro Profiling and Mass Balance of the Anti-Cancer Agent Laromustine [14C]-VNP40101M by Rat, Dog, Monkey and Human Liver Microsomes
A-E. F. Nassar* , J. Du,, M. Belcourt, X. Lin , I. King
Abstract
Laromustine (VNP40101M; 1,2-bis(methylsulfonyl)-1-(2-chloroethyl)-2-(methylamino) carbonylhydrazine) is a novel sulfonylhydrazine alkylating agent. For the first time In vitro profiling and mass balance of [14C] VNP40101M in rat, dog, monkey and human liver microsomes was investigated. Also, the role of human cytochrome P450 (CYP) and flavin-containing monooxygenase (FMO) enzymes in the conversion of [14C] VNP40101M by NADPH-fortified human liver microsomes was determined. In this study, [14C]-VNP40101M was converted to five radioactive components (C-1, C-2, C-3, C-4 and C-7) after 60 min of incubation with dog, monkey and human liver microsomes. With the exception of C-3, the same components were detected with rat liver microsomes. In the presence of NADPH, after 60 min of incubation, the loss of substrate for rat, dog, monkey and human was 63, 82, 76 and 64%, respectively and mass balance ranged from 91.0 – 99.3%. In the absence of NADPH, after 60 min of incubation with [14C] VNP40101M (100 µM), the loss of substrate for rat, dog, monkey and human liver microsomes was 59, 53, 61 and 59%, respectively and mass balance ranged from 100.6 – 116.4%. The profiles of metabolites were similar. The relative abundance of individual metabolites was not species dependent. The formation of C-7 was not observed in zero cofactor (no NADPH) or zero-protein samples, suggesting that its formation was enzymatic. The formation of C-1, C-2, C-3, and C-4 increased with respect to incubation time. Using a panel of CYP enzymes including rCYP1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6 and 3A4, it was shown that C-7 formation was catalyzed by CYP2B6 and CYP3A4/5. The results of this study suggest that (1) P450 plays a role in C-7 formation but plays little or no role in the conversion of [14C] VNP40101M to C-1 through C-4, and (2) the relative abundance of individual degradation/metabolite products were not species dependent. These findings provide a comprehensive understanding of the metabolism of this new agent.
Article Information
Identifiers and Pagination:
Year: 2010Volume: 4
First Page: 1
Last Page: 9
Publisher Id: TODMJ-4-1
DOI: 10.2174/1874073101004010001
Article History:
Received Date: 17/11/2009Revision Received Date: 15/12/2009
Acceptance Date: 3/1/2010
Electronic publication date: 27/1/2010
Collection year: 2010
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/ which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Vion Pharmaceuticals, Inc., Four Science Park, New Haven, CT 06511, USA; Tel: 203-672-4561; Fax: 203-781-8090; E-mail: nassaral@aol.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 17-11-2009 |
Original Manuscript | In Vitro Profiling and Mass Balance of the Anti-Cancer Agent Laromustine [14C]-VNP40101M by Rat, Dog, Monkey and Human Liver Microsomes | |
INTRODUCTION
Laromustine (VNP40101M; 1,2-bis(methylsulfonyl)-1-(2-chloroethyl)-2-(methylamino)carbonylhydrazine) is an active member of a relatively new class of sulfonylhydrazine prodrugs under development as antineoplastic alkylating agents [1]. Laromustine generates two distinct types of reactive intermediates: 90CE and methyl isocyanate. 90CE generates hard chloroethylating (DNA-reactive) species that alkylate DNA at the O6-position of guanine residues and rearrange to form G-C interstrand crosslinks [1-3]. Methyl isocyanate, a soft electrophilic carbamoylating agent, binds preferentially and stoichiometrically to sulfhydryl groups and inhibits a number of enzymes, including O6-alkylguanine-DNA-alkyltransferase (AGT), a DNA-repair enzyme. Methyl isocyanate has been shown to augment the cytotoxic effect of laromustine in in vitro studies by enhancing DNA-cross link formation and apoptosis in cell lines exposed to 90CE [1]. Chemical structures of (A) laromustine (VNP40101M) and (B) VNP40107 (C-7) are shown in Fig. (1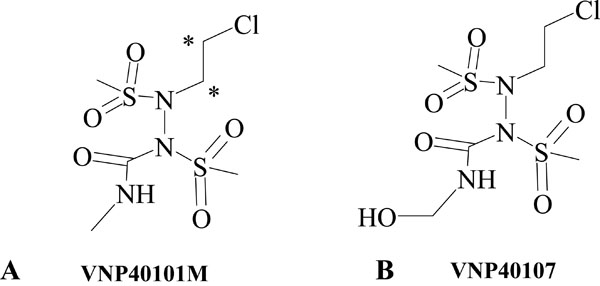 ). The average human plasma Cmax for laromustine is approximately 25 μM [4].
). The average human plasma Cmax for laromustine is approximately 25 μM [4].
 |
Fig. (1) Chemical structures of (A) laromustine (VNP40101M) and (B) VNP40107 (C-7). |
The data generated from in vitro profiling and mass balance are critical in planning and interpretation of drug safety studies. The use of in vitro comparative studies, using tissues or enzymes from humans and experimental species, it is possible to better extrapolate the risk assessment information obtained in the toxicology studies to the patient [5-7]. Reaction phenotyping (enzyme identification) studies are conducted to identify the specific enzymes responsible for the metabolism of a drug. In vitro P450 phenotyping has proven to be very successful in predicting the potential of drug interactions and the polymorphic impact on drug disposition [8-4]. To characterize the metabolic clearance of a drug candidate, the relative contribution of CYP450 enzymes to the overall elimination process is required along with identification of the enzymes responsible for the oxidative reactions. Reaction phenotyping generally involves three types of analysis: correlation analysis, antibody and chemical inhibition, and metabolism by recombinant human CYP enzymes. Each has its advantages and disadvantages, and a combination of approaches is highly recommended.
Recently we reported, an in vitro evaluation of the victim and perpetrator potential of the anti-cancer agent laromustine [4]. In the present communication, we report the in vitro metabolite profiling and mass balance of [14C]VNP40101M in rat, dog, monkey and human liver microsomes. This study showed that there is no significance differences between species in the in vitro decomposition and metabolism of [14C]-VNP40101M. Cytochrome P450 3A4 and 2B6 play a role in C-7 formation but cytochrome P450s are not involved in the conversion of [14C];VNP40101M to C-1 through C-4.
MATERIALS AND METHODS
Chemicals
Ammonium acetate, dimethyl sulfoxide (DMSO), ethylene-diaminetetraacetic acid tetrasodium salt (EDTA), glucose-6-phosphate (G6P), glucose-6-phosphate dehydrogenase (G6PDH), magnesium chloride, nicotinamide adenine dinucleotide phosphate (NADP), sodium dodecyl sulfate (SDS), sucrose and urea were purchased from Sigma-Aldrich Corporation (St. Louis, MO). Formic acid was purchased from EM Science (Gibbstown, NJ). Potassium phosphate monobasic was purchased from J.T. Baker (Phillipsburg, NJ). Methanol, acetonitrile and perchloric acid were purchased from Fisher Scientific (Pittsburgh, NJ). [14C]-VNP40101M, received from Moravek Biochemicals, Inc. EconoSafe™ was purchased from Research Products International Corporation (Mount Prospect, IL). Ultima Flo Scintillation fluid was purchased from Packard Biosciences (Boston, MA). High purity water was provided and all other reagents and solvents were of analytical grade.
Test System
Sprague-Dawley rat liver microsomes from a pool of 400, male (catalog no. R1000, lot no. 0710250, XenoTech, LLC, Lenexa, KS), Beagle dog liver microsomes from a pool of 14, male (catalog no. D1000, lot no. 0610399, XenoTech, LLC, Lenexa, KS), Cynomolgus monkey liver microsomes from a pool of 15, male (catalog no. P2000, lot no. 0610383, XenoTech, LLC, Lenexa, KS) and human liver microsomes from a pool of 50, mixed gender (catalog no. H0610, lot no. 0610351, XenoTech, LLC, Lenexa, KS) were used for this study. These microsomes were characterized with respect to the activities of various CYP enzymes. Reaction Phenotyping Kit version 7 (16 samples of human liver microsomes) were used for this study (XenoTech, LLC, Lenexa, KS).These microsomes were prepared at XenoTech and characterized with respect to the activities of various CYP enzymes. Recombinant human CYP enzymes (bactosomes) expressed in Escherichia coli, membrane protein from Escherichia coli containing empty expression plasmid and human NADPH-cytochrome c reductase were obtained from Cypex, Ltd. (Scotland, UK). The Bactosomes were characterized by the manufacturer with respect to the activities of various CYP enzymes. Recombinant human CYP enzymes co-expressed with cytochrome b5 (Supersomes), recombinant human FMO3 enzymes and the corresponding control microsomes (from non-transfected insect cells co-expressed with cytochrome b5) were purchased from BD-Biosciences (Woburn, MA). The CYP and FMO3 Supersomes were characterized by the manufacturer with respect to the activities of CYP and FMO3 enzymes.
HPLC METHOD SETUP AND PRELIMINARY EVALUATIONS
HPLC Method Setup
An HPLC method with radiometric detection was used for the detection and separation of radioactive components from [14C]-VNP40101M. The analysis was carried out with a Shimadzu LC-10AVP HPLC system, which consisted of a SIL-10A autosampler (Shimadzu Scientific Instruments, Columbia, MD) and a β;RAM radiometric detector (IN/US Systems Inc., Tampa, FL). To study the metabolism of [14C]-VNP40101M to C-7, sample analysis was carried out using an HPLC system with equipped with a radioactivity detector and consisted of two Shimadzu LC-10ADvp pumps, a Shimadzu auto sampler, and a Shimadzu DGU-14A degasser (Columbia, MD) succeeded by an IN/US Systems (-Ram radioactivity detector with a 500 ìL UHP LiGl flow cell (Tampa, FL).
[14C]-VNP40101M and its radioactive components were resolved and monitored by a gradient reversed-phase method. The column used was a Prodigy C18, 5 µm particle size, 250 ( 2.0 mm column (Phenomenex Torrance, CA), which was preceded by a direct connection guard column with C8, 4.0 mm ( 2.0 mm cartridge (Phenomenex Torrance, CA). The column was maintained at 40 ± 5°C with a column heater, and the flow rate was 0.15 mL/min from 0;30 min and 0.25 mL/min from 30-60 min. The HPLC gradient program was executed over a period of 60.0 min with water containing 5.0 mM ammonium acetate (mobile phase A) and methanol/0.1% formic acid (mobile phase B). A linear gradient used increasing proportions of 0.1% formic acid in methanol and aqueous ammonium acetate. The decomposition/metabolite products eluted between 5 and 42 min with adequate separation efficiency. Both [14C]-VNP40101M and C-7 were quantified with a calibration curve constructed using the radioactivity response of standard solutions of [14C]-VNP40101M.
Establishment of Reaction Parameters
An initial experiment was performed to establish appropriate incubation conditions leading to ~50% loss of the parent compound with liver microsomes from rat, dog, monkey and human. The purpose of achieving 50% loss of parent was to observe sufficient metabolism and/or decomposition of the parent. Liver microsomes (1 mg protein/mL) were incubated in duplicate with [14C];VNP40101M (100 µM, 2.85 µCi/incubation) at 37 ± 1°C in 0.5 mL incubation mixtures containing potassium phosphate buffer (50 mM, pH 7.4), MgCl2 (3 mM) and EDTA (1 mM, pH 7.4) with an NADPH-generating system for 0, 15, 30, 45 and 60 min. The NADPH-generating system consisted of NADP (1 mM, pH 7.4), glucose-6-phosphate (5 mM, pH 7.4) and glucose-6-phosphate dehydrogenase (1 Unit/mL). [14C];VNP40101M was added to each incubation in 0.1%(v/v) of DMSO; the concentration was based on the specific activity of 57 mCi/mmol. Reactions were initiated by the addition of [14C]-VNP40101M and were terminated at multiple time points by the addition of 500 µL of 20%(v/v) perchloric acid. Precipitated protein was removed by centrifugation at 920 × g for 10 min at 10(C. Zero-time incubations served as blanks. The supernatant fractions20 µLwere analyzed, by HPLC with Beta-RAM detection. In addition, an aliquot (100 µL) of the supernatant fractions from the stopped incubation samples was counted by liquid scintillation counting (Packard TriCarb® Scintillation Analyzer [Packard Instrument, Meriden, CT]) to determine radioactivity and percent recovery. After removal of the supernatant fraction, protein pellets were solubilized in 1 mL of 4 M urea containing 1%(w/v) SDS and were added to scintillation fluid to determine the amount of radioactivity by liquid scintillation counting.
Another preliminary experiment was designed to establish initial rate conditions (conditions where metabolite formation was proportional with respect to incubation time and protein concentration, and substrate loss was less than 20%, ideally less than 10%) for the formation of metabolite, C-7. Briefly, [14C];VNP40101M (25 and 100 µM) was incubated with pooled human liver microsomes (1 mg protein/mL) in 0.25-mL incubation mixtures containing phosphate buffer (50 mM, pH 7.4), MgCl2 (3 mM), EDTA (1 mM, pH 7.4) and an NADPH-generating system. Incubation periods were 0, 5, 10, 15 and 30 min. Reactions were initiated by the addition of [14C]-VNP40101M and were terminated by the addition of stop reagent (0.25 mL of 20% v/v perchloric acid). Incubations were performed in duplicate and were carried out at 37 ± 1oC. Precipitated protein was removed by centrifugation (920 x g for 10 min at 10°C). The supernatant fractions were analyzed with an HPLC method equipped with radiometric detection as already described. Calibration standards of [14C];VNP40101M were employed to quantify formation of C-7 and to quantify loss ofparent compound based on radiometric response. Zero-time, zero-substrate, zero-protein and zero-cofactor incubations served as controls.
REACTION PHENOTYPING
Recombinant Human CYP and FMO Enzymes
This experiment was conducted to assess the involvement of individual recombinant human CYP and flavin monooxygenase enzymes (namely FMO3) in the metabolism of [14C];VNP40101M. Briefly, [14C];VNP40101M (25 and 100 μM) was incubated in duplicate with a panel of recombinant human CYP enzymes (rCYP1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6 and 3A4) at 25 pmol P450/incubation and recombinant FMO3 at 1 mg protein/mL for 10 min at 37 ± 1oC. Recombinant CYP enzymes obtained from BD-Biosciences (rCYP2A6, rCYP2C8, rCYP2C9, rCYP2C19, and rCYP3A4) were co-expressed with cytochrome b5. All other recombinant CYP enzymes were obtained from Cypex Ltd. and were not co-expressed with cytochrome b5. FMO-3 was obtained from BD-Biosciences. Calibration standards of [14C];VNP40101M were employed to quantify formation of metabolite C-7 and to quantify loss of parent compound based on radiometric response. Zero-time and zero-protein incubations served as controls.
Correlation Analysis
[14C]-VNP40101M (25 µM) was incubated with a bank of human liver microsomes (n = 16) to determine the inter-individual differences in metabolite (C-7) formation and substrate disappearance. The following individual samples of human liver microsomes were used: numbers 196, 245, 258, 259, 273, 276, 277, 287, 288, 290, 292, 295, 297, 298, 300 and 305. The 16 samples of human liver microsomes used in this experiment were characterized at the Testing Facility (Xenotech) to determine the sample-to-sample variation in the activity of several CYP enzymes (namely CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4/5 and CYP4A11) and FMO. Differences in the rate of formation of C-7 to [14C]-VNP40101M were compared with the sample-to-sample variation on the following activities: CYP1A2 (phenacetin O-dealkylase), CYP2A6 (Coumarin 7-hydroxylase), CYP2B6 (Bupropion hydroxylase), CYP2C8(Paclitaxel 6α-hydroxylase), CYP2C9 (Diclofenac 4´-hydroxylase), CYP2C19 (S-Mephenytoin 4´-hydroxylase), CYP2D6 (Dextromethorphan O-demethylase), CYP2E1(Chlorzoxazone 6-hydroxylase), CYP3A4/5 (Testosterone 6β-hydroxylase and Midazolam 1´;hydroxylase), CYP4A11 (Lauric acid 12-hydroxylase), FMO (Benzydamine N-oxidase). Briefly, [14C]-VNP40101M (25 µM) was incubated in duplicate for zero and 10 min at 37 ± 1°C with a single concentration of human liver microsomes (1 mg protein/mL) in 0.25-mL.
Data Processing
Data were processed with the spreadsheet computer program Microsoft Excel (Microsoft Excel 2003, Microsoft Corp., Redmond, WA). To quantify C-7 formation, calibration standards of the parent were employed to calculate concentration based on radiometric response (calibration curves included two replicates of six calibration standards) using Analyst 1.4.1 or higher MS System software. Unchanged amount of [14C];VNP40101M was quantified with calibration standards of the parent compound to determine percent loss of substrate. Correlation analysis was performed with the computer software SigmaStat (Version 2.03, Systat Software Inc., Richmond, CA). Graphs were prepared using the computer software program, Microsoft Excel (Microsoft Excel 2003, Microsoft Corp., Redmond, WA).
Additional Controls
In each reaction phenotyping experiment, additional incubations were conducted at half and twice the normal protein concentration (1 mg protein/mL) and half and twice the normal incubation period (10 min) to ascertain whether metabolite formation was directly proportional to protein concentration and incubation time from experiment to experiment. Production of NADPH by the NADPH-generating system was verified spectrophotometrically.
RESULTS AND DISCUSSION
In Vitro Profiling of [14C]-VNP40101M by Rat, Dog, Monkey and Human Liver Microsomes
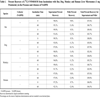
Results from the evaluation of the in vitro profile of [14C];VNP40101M by rat, dog, monkey and human liver microsomes in the presence and absence of NADPH are summarized in Tables 1, 2 and 3. Five radioactive compo-nents (C-1, C-2, C-3, C-4 and C-7) were detected after 60 min of incubation. Chromatogram of [14C]-laromustine (100 µM) with rat human liver microsomes in the presence of NADPH is shown in Fig. (2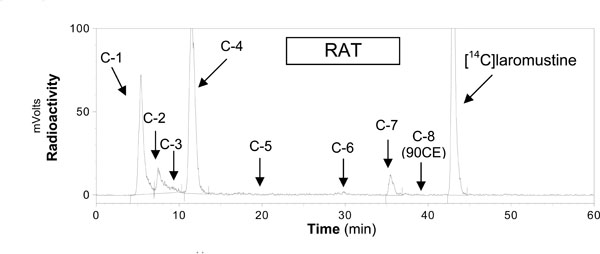 ). [14C];VNP40101M (100 µM) was extensively metabolized and/or degraded in both the presence and absence of NADPH. C-7 was the only component whose formation was dependent on NADPH. Formation of the five components accounted for the loss of substrate as shown in Table 3. In the presence of NADPH, after 60 min of incubation, the loss of substrate for rat, dog, monkey and human was 63, 82, 76 and 64%, respectively and mass balance ranged from 91.0 – 99.3%. In the absence of NADPH, after 60 min of incubation with [14C];VNP40101M (100 µM), the loss of substrate for rat, dog, monkey and human liver microsomes was 59, 53, 61 and 59%, respectively and mass balance ranged from 100.6 – 116.4%. For rat, dog, monkey and human, the total formation (sum of C-1, C-2, C-3, C-4 and C-7) was 54, 81, 70 and 55%, respectively in the presence of NADPH and 59, 69, 66 and 64%, respectively, in the absence of NADPH. Formation of C-7 did not account for the loss of parent compound. The loss of substrate was largely caused by non-enzymatic chemical degradation. In contrast, formation of C-7 was not observed in zero;cofactor (no NADPH) and zero-protein samples suggesting its formation was enzymatic.
). [14C];VNP40101M (100 µM) was extensively metabolized and/or degraded in both the presence and absence of NADPH. C-7 was the only component whose formation was dependent on NADPH. Formation of the five components accounted for the loss of substrate as shown in Table 3. In the presence of NADPH, after 60 min of incubation, the loss of substrate for rat, dog, monkey and human was 63, 82, 76 and 64%, respectively and mass balance ranged from 91.0 – 99.3%. In the absence of NADPH, after 60 min of incubation with [14C];VNP40101M (100 µM), the loss of substrate for rat, dog, monkey and human liver microsomes was 59, 53, 61 and 59%, respectively and mass balance ranged from 100.6 – 116.4%. For rat, dog, monkey and human, the total formation (sum of C-1, C-2, C-3, C-4 and C-7) was 54, 81, 70 and 55%, respectively in the presence of NADPH and 59, 69, 66 and 64%, respectively, in the absence of NADPH. Formation of C-7 did not account for the loss of parent compound. The loss of substrate was largely caused by non-enzymatic chemical degradation. In contrast, formation of C-7 was not observed in zero;cofactor (no NADPH) and zero-protein samples suggesting its formation was enzymatic.
 |
Fig. (2) Chromatogram from incubations of [14C]-laromustine (100 µM) with rat human liver microsomes in the presence of NADPH. |
In vitro studies were designed to determine the role of P450 and other HLM enzymes on VNP40101M metabolism. Incubations of [14C];VNP40101M were performed with and without human liver microsomes and with boiled (denatured) microsomes, with and without NADPH. The data suggested that [14C];VNP40101M undergoes extensive chemical degradation rather than metabolism by cytochrome P450s. It was also determined that reactions stopped with methanol, acetonitrile and perchloric acid generated similar degradation profiles. These results suggest that the sample preparation methods did not contributed to the degradation products.
IDENTIFICATION OF HUMAN CYP ENZYMES INVOLVED IN THE METABOLISM OF [14C]-VNP40101M: REACTION PHENOTYPING
Recombinant Human CYP and FMO Enzymes
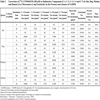
[14C]-VNP40101M (25 and 100 µM) was incubated with a panel of recombinant human CYP enzymes (rCYP1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6 and 3A4) and recombinant human FMO3 for zero and 10 min. Results are summarized in Tables 4, 5 and 6 and Figs. (3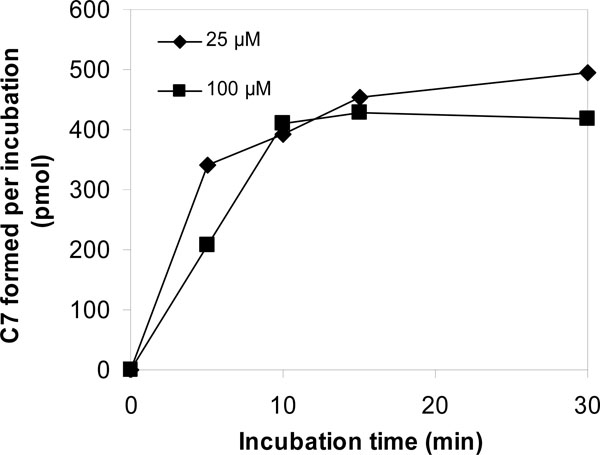 and 4
and 4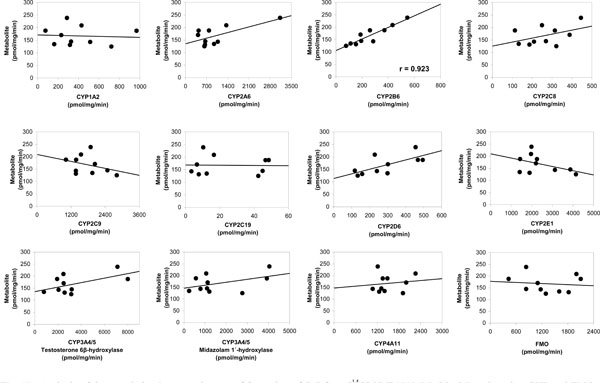 ). Among the panel of recombinant human CYP enzymes and FMO enzyme evaluated, formation of C-7 was only observed in incubations with recombinant CYP2B6 and CYP3A4. Loss of parent compound was observed in all incubations including thecontrol samples (i.e., samples containing membranes with no human CYP enzyme). Formation of C-7 was observed in incubations containing recombinant human CYP2B6 and CYP3A4 at both substrate concentrations. After 10;min incubations of [14C];VNP40101M at concentrations 25 and 100 µM with recombinant human CYP2B6, the [14C] amounts of C-7 were 376 and 513 pmol, respectively. After 10-min incubations of [14C];VNP40101M at concentrations 25 and 100 µM with recombinant human CYP3A4, the [14C] amounts of C-7 were 362 and 549 pmol, respectively.The formation of the radioactive components increased with respect to incubation time with the exception of C-7. The formation of C-7 tended to increase with increasing incubation time and protein concentration; however, formation of C-7 was not linear with respect to either of these parameters. In addition, the formation of C-7 was not proportional to substrate concentration. The non-linearity of C-7 formation suggests that, once formed, this component was further metabolized or degraded.
). Among the panel of recombinant human CYP enzymes and FMO enzyme evaluated, formation of C-7 was only observed in incubations with recombinant CYP2B6 and CYP3A4. Loss of parent compound was observed in all incubations including thecontrol samples (i.e., samples containing membranes with no human CYP enzyme). Formation of C-7 was observed in incubations containing recombinant human CYP2B6 and CYP3A4 at both substrate concentrations. After 10;min incubations of [14C];VNP40101M at concentrations 25 and 100 µM with recombinant human CYP2B6, the [14C] amounts of C-7 were 376 and 513 pmol, respectively. After 10-min incubations of [14C];VNP40101M at concentrations 25 and 100 µM with recombinant human CYP3A4, the [14C] amounts of C-7 were 362 and 549 pmol, respectively.The formation of the radioactive components increased with respect to incubation time with the exception of C-7. The formation of C-7 tended to increase with increasing incubation time and protein concentration; however, formation of C-7 was not linear with respect to either of these parameters. In addition, the formation of C-7 was not proportional to substrate concentration. The non-linearity of C-7 formation suggests that, once formed, this component was further metabolized or degraded.
 |
Fig. (3) Effect of incubation time and substrate concentration on the conversion of [14C]-VNP40101M (25 and 100 100 µM) to C-7 by NADPH-fortified human liver microsomes. |
Correlation Analysis
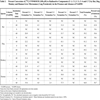
Results are shown in Table 6 and Fig. (4 ). The sample-to-sample variation in the formation of C-7 from [14C]-VNP40101M (25 µM e.g. 0.25 µM (2.5 pmol per injection)) was evaluated among 16 individual samples. The formation of C-7 was not observed in four individuals all of which had low CYP2B6 activity. The data suggested that two individuals were outliers for CYP2C19 activity resulting in an artificially high correlation to CYP2C19 activity. Therefore, these two individuals were not included. The sample-to-sample variation (n = 10) in the formation of C-7 correlated strongly with CYP2B6 activity as measured by bupropion hydroxylation (r = 0.923). Formation of C-7 correlated to a lesser extent (i.e., r = 0.5 or greater) with three other CYP activities, namely CYP2A6 activity as measured by coumarin 7;hydroxylation (r = 0.678), CYP2D6 activity as measured by dextromethorphan O-demethylation(r = 0.691) and CYP3A4 activity as measured by testosterone 6β-hydroxyl-ation (r = 0.572). Although the correlation with CYP2A6 and CYP2D6 activity were moderate, it was possible that the correlation observed was an artifact due to the enzyme-to-enzyme correlation with CYP2B6 activity among the individual microsomal samples (i.e., r = 0.598 and r = 0.401).
). The sample-to-sample variation in the formation of C-7 from [14C]-VNP40101M (25 µM e.g. 0.25 µM (2.5 pmol per injection)) was evaluated among 16 individual samples. The formation of C-7 was not observed in four individuals all of which had low CYP2B6 activity. The data suggested that two individuals were outliers for CYP2C19 activity resulting in an artificially high correlation to CYP2C19 activity. Therefore, these two individuals were not included. The sample-to-sample variation (n = 10) in the formation of C-7 correlated strongly with CYP2B6 activity as measured by bupropion hydroxylation (r = 0.923). Formation of C-7 correlated to a lesser extent (i.e., r = 0.5 or greater) with three other CYP activities, namely CYP2A6 activity as measured by coumarin 7;hydroxylation (r = 0.678), CYP2D6 activity as measured by dextromethorphan O-demethylation(r = 0.691) and CYP3A4 activity as measured by testosterone 6β-hydroxyl-ation (r = 0.572). Although the correlation with CYP2A6 and CYP2D6 activity were moderate, it was possible that the correlation observed was an artifact due to the enzyme-to-enzyme correlation with CYP2B6 activity among the individual microsomal samples (i.e., r = 0.598 and r = 0.401).
The linear regression lines for the aforementioned enzymes did not pass through or near the origin, which suggests that multiple enzymes are involved in the formation of C-7. Multivariate regression analysis was performed with CYP2B6 and CYP3A4 to further assess the contribution of these enzymes to the formation of C;7. This approach can successfully identify enzymes involved when each enzyme contributes 25% or more to metabolite formation, but it will likely not identify an enzyme that contributes substantially less than 25% [15]. The sample-to-sample variation in the formation of C-7 correlated well with CYP2B6 activity (r = 0.923), but the correlation improved when the variation in CYP3A4 activity (as measured by testosterone hydroxylation) was taken into consideration (r = 0.945).
Additional Controls
In each reaction phenotyping experiment, additional incubations were conducted at half and twice the normal protein concentration (1 mg protein/mL) and half and twice the normal incubation period (10 min) to ascertain whether metabolite formation was directly proportional to protein concentration and incubation time from experiment to experiment. In incubations with NADPH-fortified pooled human liver microsomes, the formation of C;7 tended to increase with increasing incubation time and protein concentration; however, formation of C-7 was not linear with respect to either of these parameters. In addition, the formation of C-7 was not proportional to substrate concentration.
CONCLUSION
Liver microsomes from dog, monkey and human converted [14C];VNP40101M to five components (C-1, C-2, C-3, C-4 and C-7), four of which (all but C-3) were also formed by rat liver microsomes. Either C-3 was not formed or below the detection limit in rat liver microsomes. Only the formation of C-7 required the presence of NADPH. C-7 appears to be decomposed or metabolized further. These results suggest that cytochrome P450 plays a role in C-7 formation but plays little or no role in the conversion of [14C]-VNP40101M to C-1 through C-4. The combined results from the correlation analysis and recombinant human CYP enzymes experiments point to a role for both CYP2B6 and CYP3A4 in the formation of metabolite C-7. The formation of C-7 correlated the strongest with CYP2B6 activity with moderate correlation to CYP2A6, CYP2D6 and CYP3A4 activity in 10 individual samples of NADPH-fortified human liver microsomes. As shown by the recombinant human CYP data, rCYP3A4 and rCYP2B6 were capable of forming C-7, whereas formation of C-7 was not observed with rCYP2A6 and rCYP2D6. VNP40101M was shown to be a competitive inhibitor for both CYP2B6 and CYP3A4, which is consistent with VNP40101M being a substrate for these enzymes (Nassar et al., 2009 DMD). Combined, these findings provide an understanding of the metabolism of this new agent. The identification of the decomposition/metabolic products and in vitro phase II studies of laromustine are on-going.