- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Medical Informatics Journal
(Discontinued)
ISSN: 1874-4311 ― Volume 13, 2019
Prototypes for Content-Based Image Retrieval in Clinical Practice
Adrien Depeursinge*, 1, 2, Benedikt Fischer 3, Henning Müller 1, 2, Thomas M Deserno 3
Abstract
Content-based image retrieval (CBIR) has been proposed as key technology for computer-aided diagnostics (CAD). This paper reviews the state of the art and future challenges in CBIR for CAD applied to clinical practice.
We define applicability to clinical practice by having recently demonstrated the CBIR system on one of the CAD demonstration workshops held at international conferences, such as SPIE Medical Imaging, CARS, SIIM, RSNA, and IEEE ISBI. From 2009 to 2011, the programs of CADdemo@CARS and the CAD Demonstration Workshop at SPIE Medical Imaging were sought for the key word “retrieval” in the title. The systems identified were analyzed and compared according to the hierarchy of gaps for CBIR systems.
In total, 70 software demonstrations were analyzed. 5 systems were identified meeting the criterions. The fields of application are (i) bone age assessment, (ii) bone fractures, (iii) interstitial lung diseases, and (iv) mammography. Bridging the particular gaps of semantics, feature extraction, feature structure, and evaluation have been addressed most frequently.
In specific application domains, CBIR technology is available for clinical practice. While system development has mainly focused on bridging content and feature gaps, performance and usability have become increasingly important. The evaluation must be based on a larger set of reference data, and workflow integration must be achieved before CBIR-CAD is really established in clinical practice.
Article Information
Identifiers and Pagination:
Year: 2011Volume: 5
Issue: Suppl 1
First Page: 58
Last Page: 72
Publisher Id: TOMINFOJ-5-58
DOI: 10.2174/1874431101105010058
Article History:
Received Date: 16/5/2011Revision Received Date: 20/5/2011
Acceptance Date: 20/5/2011
Electronic publication date: 27/7/2011
Collection year: 2011
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Business Information Systems, University of Applied Sciences Western Switzerland (HES–SO), TechnoArk 3, 3960 Sierre, Switzerland; Tel +41 27 606 9023; Fax +41 27 606 9000; E-mail: adrien.depeursinge@hevs.ch
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 16-5-2011 |
Original Manuscript | Prototypes for Content-Based Image Retrieval in Clinical Practice | |
1. . INTRODUCTION
1.1. History of Medical CBIR
In the early 1990s, the query by image content (QBIC) system of IBM was one of the first approaches to content-based image retrieval (CBIR), and the query by image example (QBE) paradigm has since been established [1Niblack W, Barber R, Equitz W, et al. The QBIC project: querying images by content using color, texture and shape Proc SPIE 1993; 1908: 173-87.]. Representing images by means of numerical features (signature), relevant images are identified by comparing the signature of an example with all signatures in a repository. Initially, CBIR was applied to images from the Internet or large volumes of photographs [2Smeulders AWM, Worring M, Santini S, Gupta A, Jain R. Content-based image retrieval at the end of the early years IEEE Trans Pattern Anal Mach Intell 2000; 22(12): 1349-80.]. The signatures were obtained from color, texture, and shape. Since color has been identified as the most relevant structure for CBIR, the semantic gap was recognized. It describes the differences between image similarity on the high level of human perception and the low level of a few numerical numbers describing a mean color.
A comprehensive review of CBIR systems in medical applications is given by Müller et al. [3Müller H, Michoux N, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medical applications: clinical benefits and future directions Int J Med Inform 2004; 73(1): 1-23.]. To narrow the semantic gap, first medical computer-aided diagnosis- (CAD) CBIR approaches focused on a particular modality and application domain, such as microscopy, photography of the skin, radiographs of spine, teeth, and mamma, computed tomography (CT) of lungs, and magnetic resonance imaging (MRI) of the head (Table 1). According to Tagare et al., medical image information further contains spatial data, and a large part of image information is geometric [4Tagare HD, Jaffe CC, Duncan J. Medical image databases: a content-based retrieval approach J Am Med Inform Assoc 1997; 4(3): 184-98.]. Accordingly, initial attempts in bridging the semantic gap were based also on local image signatures referring to pre-segmented regions of interest (ROI) and relative positions of relevant objects (Table 1).
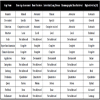
Gap Classification of Prototypes According to [20Deserno TM, Antani S, Long R. Ontology of gaps in content-based image retrieval J Digit Imaging 2009; 22(2): 202-15.]
With the automatic search and selection engine with retrieval tools (ASSERT) system, for instance, the physician manually delineates the pathology bearing ROI and a set of anatomical landmarks when an image is entered into the database [5Shyu CR, Brodley CE, Kak AC. ASSERT: a physician-in-the-loop content-based retrieval system for HRCT image databases Comput Vis Image Underst 1999; 75(1-2): 111-32.]. However, driven by the ever-increasing amount of medical image data acquired directly in the digital form in today’s clinical practice, manual annotations are time consuming, imprecise, irreproducible, and simply impracticable.
The development of rather general approaches such as I-Browse and KMeD began about ten years ago. The established frameworks for medical CBIR systems today are the medical GNU image finding tool (MedGIFT) and the image retrieval in medical applications (IRMA) project [15Lehmann TM, Güld MO, Thies C, et al. Content-based image retrieval in medical applications Methods Inf Med 2004; 43(4): 354-61.,16Güld MO, Thies C, Fischer B, Lehmann TM. A generic concept for the implementation of medical image retrieval systems Int J Med Inform 2007; 76(2-3): 252-9.], which started in 2002 and 2001, respectively.
1.2. Fields of Engineering
Simple CBIR prototypes can be developed quickly by computer scientists from the fields of image processing and machine learning, being central to the characterization of image content. To build large-scale CBIR systems however, expertise from several fields of engineering must be combined to fulfil the needs of specialized user groups including a high level of interaction (Table 2) [2Smeulders AWM, Worring M, Santini S, Gupta A, Jain R. Content-based image retrieval at the end of the early years IEEE Trans Pattern Anal Mach Intell 2000; 22(12): 1349-80.]. Initially, raw, low-level information is extracted from original images using image processing techniques. These features are usually characterizing color, shape and texture, either globally or within an image region (locally). Then, computer vision, machine learning as well as knowledge from human vision and psychology are required to aggregate, optimize and map the low-level features to high-level semantic concepts driven by user’s intents and visual perception of the image collection of concern [17Zhou XS, Zillner S, Moeller M, et al. Semantics and CBIR: a medical imaging perspective International Conference on Content- Based Image and Video Retrieval, Niagara Falls, Canada 2008 2008; 571-80.]. Another important cue is to maximize human-computer interaction with appropriate interfaces for query formulation and result visualization. In particular, allowing physicians to efficiently draw a sketch or quickly mark a volume of interest in 3D or in multiple-captured volumes (3D+time) is still an insufficiently solved problem. Studies in information retrieval also showed that involving the user in the loop for query refinement enables quicker convergence between user’s intents and retrieved results [18Rui Y, Huang TS, Ortega M, Mehrotra S. Relevance feedback: a power tool for interactive content-based image retrieval IEEE Trans Circ Syst Video Technol 1998; 8(5): 644-55.]. On the technical side, efficient database management and high-performance computing are both required to optimize the retrieval quality offline (e.g., parameter optimization over large image collections) and to ensure quick online system response [19Costa Oliveira M, Walfredo C, de Azevedo Marques PM. Towards applying content-based image retrieval in the clinical routine Future Gen Comput Syst 2007; 23(3): 466-74.].
1.3. CBIR vs CAD
The most popular approach to image-based CAD aims at providing automated interpretation of image examinations as a second opinion to radiologists. These systems have proved to be particularly useful for analysing large amounts of data containing easily detectable lesions but with low prevalence [22Doi K. Computer-aided diagnosis in medical imaging: historical review, current status and future potential Comput Med Imaging Graph 2007; 31(4-5): 198-211.]. When compared to human readers, computers can efficiently and exhaustively analyse large numbers of images with high reproducibility. In the literature, two types of CAD systems are distinguished:
- Computer-aided detection (CADe) aims at pre-analyzing images and automatically annotates suspicious regions in order to support the radiologist in reading. No classification of the ROIs is done.
- Computer-aided diagnostics (CADx) aims at deriving a diagnostic decision by adding a classification step, where the identified ROIs are analysed, classified, and a semantic conclusion is drawn, which might serve the radiologist as second opinion.
The initial attempts to CADx were carried out more than forty years ago in chest X-ray imaging [23Meyers PH, Nice CM, Becker HC, et al. Automated computer analysis of radiographic images Radiology 1964; 83: 1029-34.,24Kruger RP, Thompson WB, Turner AF. Computer diagnosis of pneumoconiosis IEEE Trans Syst Man Cybern 1974; 4(1): 40-9.]. These systems aimed at replacing radiologists as the originators relied on the assumption that computers were better at performing certain tasks than human beings. However, it quickly became clear that physicians and radiologists have to take the final decision and the outputs of CAD systems must be used as second opinions and information providers [25Engle RL. Attempts to use computers as diagnostic aids in medical decision making: a thirty-year experience Perspect Biol Med 1992; 35(2): 207-19.]. Recently, CADe systems have been used in clinical practice in the rather mature field of cancer screening in mammograms and allowed to improve the detection of non–palpable cancerous masses [26Vomweg TW. Computer-aided diagnosis: clinical applications in the breast In: Image Processing in Radiology, Medical Radiology. Berlin: Springer 2008; pp. 355-74.]. Other notable examples are the assisted detection of lung nodules in chest radiographs such as Riverain Medical’s OnGuard1 which have obtained approval from the United States’ Food and Drug Administration (FDA). Furthermore, CADe is expected to be introduced into clinical routine for several other domains such as the chest, colon, brain, liver, skeletal and vascular systems [22Doi K. Computer-aided diagnosis in medical imaging: historical review, current status and future potential Comput Med Imaging Graph 2007; 31(4-5): 198-211.].
In summary, medical CBIR systems are well aligned with early 1990s’ conclusions that CAD should be used as second opinion and information providers [25Engle RL. Attempts to use computers as diagnostic aids in medical decision making: a thirty-year experience Perspect Biol Med 1992; 35(2): 207-19.], rather than independent automatic diagnostic systems. However, despite almost 20 years of intensive research in academia, CBIR has not reached as far as beyond research labs and, to our knowledge, no commercially available medical CBIR system exists yet. It is all the more surprising that the techniques in image processing and machine learning used for CADe and CBIR are similar in terms of structure, and the major disparities between the two occur in graphical user interfaces (GUI), clinical workflows and integration.
In this work, we try to answer the question why CBIR systems did not reach clinical practice yet. We provide a detailed analysis of CBIR systems that are close to be integrated and analyse their strengths and pitfalls. The corresponding unaddressed gaps are identified, and, from that, future directions are provided in a hope to foster the adoption of CBIR systems in clinical radiology.
2. MATERIALS AND METHODS
2.1. State of the Art in Reviewing Systems
Reviewing medical CBIR systems is an often discussed issue with the first paper appeared in 1997 [4Tagare HD, Jaffe CC, Duncan J. Medical image databases: a content-based retrieval approach J Am Med Inform Assoc 1997; 4(3): 184-98.]. Thereafter, reviews usually are specialized on a certain medical or technological application domain such as forensics [27Geradts Z, Bijhold J. Content based information retrieval in forensic image databases J Forensic Sci 2002; 47(2): 285-92.] or Web 2.0 [28Müller H. Medical multimedia retrieval 2.0 Yearb Med Inform 2008; 55-63.], respectively. Rather general reviews have been published by Müller et al. [3Müller H, Michoux N, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medical applications: clinical benefits and future directions Int J Med Inform 2004; 73(1): 1-23.] and Akgul et al. [29Akgul CB, Rubin DL, Napel S, Beaulieu CF, Greenspan H, Acar B. Content-based image retrieval in radiology: current status and future directions J Digit Imaging 2011; 24(2): 208-2.] referring to CBIR in radiology by current status, clinical benefits, and future directions. Ending up with 187 or 77 references from the two reviews, respectively, inclusion or exclusion criteria are not fully clear, which limits the impact of such work. However, a somewhat more systematic methodology for classifying CBIR systems has also bee proposed [20Deserno TM, Antani S, Long R. Ontology of gaps in content-based image retrieval J Digit Imaging 2009; 22(2): 202-15.,30Long LR, Antani S, Deserno TM, Thoma GR. Content-based image retrieval in medicine: retrospective assessment, state of the art, and future directions Int J Healthc Inform Syst Informat 2009; 4(1): 1-16.].
2.1.1. Defining the Characteristics of Medical CBIR Systems
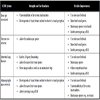
In [20Deserno TM, Antani S, Long R. Ontology of gaps in content-based image retrieval J Digit Imaging 2009; 22(2): 202-15.], for instance, a set of 14 so called gaps are defined to classify medical CBIR systems, which are enriched with additional 7 characteristics. The gaps were identified as being responsible for potential pitfalls and inadequacy of current medical CBIR systems. For instance, the “semantic gap” describes the discrepancies between a high-level of semantic in human image perception and understanding and the simple numerical signature that is extracted by a machine in terms of color, texture and shape. More systematically, the authors define four clusters of gaps:
- Content: The content gaps address the level of image understanding (1 – semantic gap) as well as the imaging and/or clinical context in which a CBIR system may be used (2 – use context gap). Obviously, designing a medical CBIR system for a broad use is more challenging, since the level of image details being relevant for the retrieval, the type of image data (modality) to handle, and other system preferences are highly variable.
- Feature: The feature gaps address the automation of feature extraction (3 – extraction gap), the granularity or dimensionality of structure of image objects recognized by the system (4 –structure gap), of visual details in the image processed by the system (5 –scale gap), of spatial and time inputs actually used to compute the signature (6 – space & time dimension gap), and of channel inputs actually used to compute the signature (7 – channel dimension gap).
- Performance: The performance gaps describe the levels of actual implementation of the system (8 – application gap), of integration into patient care information systems (9 – integration gap), of support for fast database searching (10 – indexing gap), and to which the system validity of retrieval has been evaluated (11 –evaluation gap).
-
Usability: The usability gaps address the levels to which user may use and combine text and visual queries (12 – query gap), to which the system helps the user to understand query results (13 – feedback gap), and to which the system enable the user to refine and improve query results (14 –refinement gap).
These gaps highlight the need of contributions from several fields of engineering in order to successfully design, implement, and integrate medical CBIR systems into clinical practice (Table 2). With respect to our intention, the integration gap is of particular relevance. Already in 1999, Eakins & Graham explicitly have stated with respect to the non-medical CBIR application domain that “the experience of all commercial vendors of CBIR software is that system acceptability is heavily influenced by the extent to which image retrieval capabilities can be embedded within users’ overall work tasks” [32Eakins J, Graham M. Content-based image retrieval Technical Report No 39, JISC Technology Applications Programme, Newcastle, UK: University Northumbria 1999.]. This is entirely true for medical applications as well.
2.1.2. . Defining the Review Methodology
According to previous work in general image retrieval [33Datta R, Joshi D, Li J, Wang JZ. Image retrieval: ideas, influences, and trends of the new age ACM Compute Surv 2007; 40(2): 1-60.], Long et al. proposed to formalize the review methodology to retrospectively assess the state of the art and future directions of medical CBIR systems [30Long LR, Antani S, Deserno TM, Thoma GR. Content-based image retrieval in medicine: retrospective assessment, state of the art, and future directions Int J Healthc Inform Syst Informat 2009; 4(1): 1-16.]. Different criteria were defined:
- Journals: The journals were identified using informal selection criteria, but with the goal of providing a broad representation of the major publications reporting medical image retrieval research results. Ten journals from engineering and medicine were included.
- Date: Aiming at reviewing recent work, the date of publication was set between 2001 and 2010. Defining an ending date may be useful since the processes of paper writing, reviewing, and publishing may take up to five years [34Spreckelsen C, Deserno TM, Spitzer K. The publication echo: effects of retrieving literature in PubMed by year of publication Int J Med Inform 2010; 79(4): 297-303.].
- Database: Several bibliometric databases and search tools are available. For instance, Pubmed (National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD, USA), Google Scholar (Google Corp., Mountain View, CA, USA), and the ISI Web of Science (Thomson Reuters Corp., New York, NY, USA) are most recognized. However, the visibility of medical informatics regarding may differ with respect to coverage and completeness [35Spreckelsen C, Deserno TM, Spitzer K. Visibility of medical informatics regarding bibliometric indices and databases BMC Med Inform Decis Mak 2011; 11(1): 24.].
- Terms: The search phrases must also be determined in order to produce reproducible results. In the work of Long et al., the terms [“medical image retrieval AND search_phrase] were used, where search_phrase was one of eleven CBIR-related phrases including, for instance, “content-based image retrieval], “Indexing” , “Performance” , or “Relevance Feedback” [30Long LR, Antani S, Deserno TM, Thoma GR. Content-based image retrieval in medicine: retrospective assessment, state of the art, and future directions Int J Healthc Inform Syst Informat 2009; 4(1): 1-16.].
2.1.3. Evaluations with Clinical Practitioners
Another idea to assess the “user readiness” of CBIR systems was proposed by Antani et al. [31Antani S, Xue Z, Long LR, et al. Is there a need for biomedical CBIR systems in clinical practice? outcomes from a usability study Proc SPIE 2011; 7967]. The authors applied a set of usability evaluation methods known from quantitative and qualitative research to evaluate exemplarily a CBIR system supporting the access to 100,000 cervigrams and related, anonymized patient data. These methods include:
- Questionnaire: Designed for the purpose of gathering information from respondents, a series of questions and other prompts or scales are presented either paper-based or electronically. Such questionnaires are filled by the user and usually analyzed statistically.
- Structured Interview: Similar to paper-based surveys (questionnaire), the users are presented with exactly the same questions in the same order to support data aggregation. Such interviews are also referred to as (researcher-administered survey).
- Focus Group: A group of people is asked about their perceptions, opinions, beliefs, and attitudes towards a system. The questions are asked in an interactive setting where participants are free to talk with other group members.
- Think-aloud Method: Participants are encouraged to voice their thoughts on the system as they perform given tasks, while an expert facilitator is guiding the process of the session, and comments the voice recordings.
The methods also help to identify obstacles that hamper practical use of such systems. In the above given example, for instance, the user uncovered many problems such as it was (i) challenging to obtain a clear understanding of the purpose and functionality of the tool without any training on the tool’s capabilities, (ii) difficult to discern how to properly formulate a visual query, which is a critical component in CBIR systems, and (iii) almost impossible to use the interactive drawing tools successfully [31Antani S, Xue Z, Long LR, et al. Is there a need for biomedical CBIR systems in clinical practice? outcomes from a usability study Proc SPIE 2011; 7967].
2.2. Definition for CBIR in Clinical Practice
However, complex schemes of terminology do not guarantee unambiguous location and complete inclusion of relevant work. Although a well-defined ontology is most important to science, it may still remain ambiguous to apply these terms correctly, since relevant information may be missing in the reports that are published as scientific article. The same holds for a detailed search strategy. Including or excluding systems by user evaluation studies delivers the most objective assessment of readiness for clinical practice, but is too costly regarding both, system and man power.
In order to identify content-based image retrieval systems that are really near to clinical practice, we assume they have been demonstrated in one of the recent workshops on CAD. Such workshops may be organized by:
- CARS: The Computer-Assisted Radiology and Surgery private initiative, Kuessaberg, Germany, along with the annual CARS Conference,
- RSNA: The Radiological Society of North America, Oak Brook, IL, USA, along with its annual meeting,
- SIIM: The Society for Imaging Informatics in Medicine, Leesburg, VA, USA, along with its annual meeting,
- IEEE: The Institute of Electrical and Electronics Engineering, Piscataway, NJ, USA, along with the international Symposium for Biomedical Imaging (ISBI),
- SPIE: The Society of Photo-Optical Instrumentation Engineers, Bellingham, WA, USA, along with its annual International Symposium on Medical Imaging,
- MICCAI: The Medical Image Computing and Computer Assisted Intervention Society, Minnesota, USA, along with the annual MICCAI Conference.
The workshops were analysed in the years between 2009 and 2011, where “Computer-aided Diagnosis” and “retrieval” were in the workshop and software presentation title, respectively.
2.3. Applying Criteria
Among the six targeted conferences, two have dedicated sessions for live demonstrations of CAD systems. The events are:
- CADdemo@CARS in years 2009 and 2010 (2011 had not happened yet at the time when this paper was written),
- SPIE Medical Imaging (MI) CAD workshop in years 2009, 2010 and 2011.
Other targeted conferences (i.e., RSNA, SIIM, IEEE ISBI and MICCAI) may have live software demonstrations in the context of commercial exhibitions, but these are not dedicated to CAD systems and very few technical details can be found for commercial systems.
In total, 4 systems presented in the five workshops (CADdemo@CARS 2009-2010 and SPIE MI CAD workshops 2009-2011) and containing “retrieval” in the title were analyzed. Some other CADx systems that did not contain “retrieval” in the title may contain CBIR features but were not in the main focus of the developments. At the CADdemo@CARS 2010 in Geneva, three CBIR-based CAD systems were presented:
- A platform for bone age assessment, developed by the Rheinisch-Westfälische Technische Hochschule (RWTH) in Aachen, Germany entitled “Web-based bone age assessment using case-based reasoning and content-based image retrieval”.
- Retrieval of fracture cases for operation planning developed at the University Hospitals of Geneva (HUG), Switzerland entitled “Case-based visual retrieval of fractures”.
- Analysis and retrieval of high-resolution CT (HRCT) images from patients affected with interstitial lung diseases (ILDs) developed by the HUG entitled “Content-based retrieval and analysis of HRCT images from patients with interstitial lung diseases: a comprehensive diagnostic aid framework”.
At the SPIE MI CAD workshops 2009 and 2010, two CAD systems for the characterization of breast masses in mammograms were found with “retrieval” in the title:
- “Expert-guided content–based mammographic mass retrieval system”, developed by Georgetown University Medical Center, Washington DC, USA.
- “Content-based image retrieval (CBIR) CADx system for characterization of breast masses”, developed by University of Michigan Medical Center, USA.
The collaboration between the two research groups resulted in a publication at SPIE MI 2011 [36Tao Y, Lo SC, Hadjiski L, Chan HP, Freedman MT. BI-RADS guided mammographic mass retrieval Proc SPIE 2011; 7963: 2H.], which was used for the description of the CBIR system in Section 3.1.4. These four systems are described in details in Section 3.1, compared in Section 3.2. and analyzed in terms of gap identification in Section 3.3.
3. RESULTS
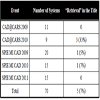
The programs of 6 conferences were analyzed. In years 2009-2011, workshops and special sessions dedicated to live demonstration of CAD systems were found in CARS and SPIE MI conferences. As CAD@CARS2011 had not happened yet at the time this paper was written, the program of five workshops (CAD@CARS 2009-2010 and SPIE MI CAD demo 2009-2011) were sought for CBIR systems. 70 CAD systems were presented, and 5 of them (7%) contained “retrieval” in the title (Table 3).
3.1. System Descriptions
3.1.1. Bone Age Assessment
With its underlying flexible structure of image processing and image retrieval algorithms, the IRMA framework was adjusted to enrich CAD in the context of bone age assessment (BAA). A live demonstration of this system was presented at the CADdemo@CARS 2010 in Geneva.
Background and Objective
Bone age assessment based on hand radiographs is a frequent and time-consuming task for radiologists to estimate the maturity of patients. Relating the bone to chronological ages and the current status of growth allows estimating adult height of pediatric subjects, as well as diagnosing and tracking endocrine disorders or pediatric syndromes [37Gilsanz V, Ratib O. Hand bone age In: A digital atlas of skeletal maturity. Berlin: Springer 2005.]. Clinically, the methods by Greulich & Pyle (GP) [38Greulich WW, Pyle SI. Radiographic Atlas of Skeletal Development of Hand Wrist Stanford University Press 1971.] or Tanner & Whitehouse (TW3) [39Tanner JM, Healy MRJ, Goldstein H, Cameron N. Assessment of skeletal maturity and prediction of adult height (TW3). London: WB Saunders 2001.] are applied. In the former method, radiologists compare all bones of the hand to those shown in radiographs from the standard atlas. In the latter case, a certain subset of bones is examined.
Several approaches have been taken to (partially) automate the BAA-process, and recently, a commercial application was reported [40Thodberg HH, Kreiborg S, Juul A, Pedersen KD. The bone xpert method for automated determination of skeletal maturity IEEE Trans Med Imaging 2009; 28(1): 52-66.]. In general, all existing approaches rely on computation and measurements of image- or region-related features, which are usually incomprehensible to the user. Providing more transparency, the IRMA-based BAA application aims at merging CBIR with case-based reasoning (CBR). This is achieved by retrieving similar radiographs with validated ages from a case database, and subsequently presenting these to a radiologist along with a suggested bone age deduced from similarity and bone age of previous cases.
Methods and Application
The development of the hand bones is most evident in certain image regions, namely the epiphyses, the carpal bones, as well as the distal radius and ulna. Therefore, ROIs are subjected to CBIR queries rather than the entire radiograph. In the current IRMA-BAA prototype, only the epiphyses are considered, while the other bones are proclaimed to be included in future releases.
For a new hand radiograph, the processing pipeline consists of four steps (Fig. 1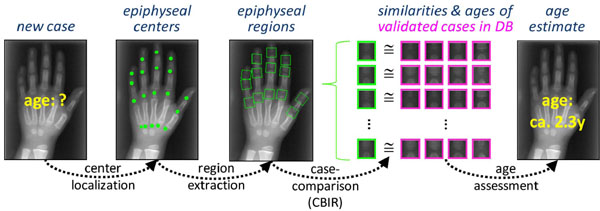 ):
):
 |
Fig. (1) CBIR approach to bone-age assessment. |
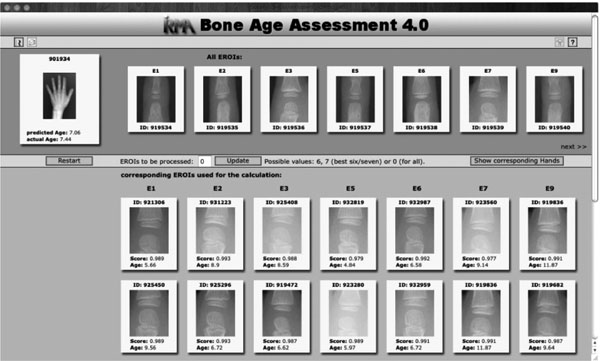 |
Fig. (2) Result display for the query image indicated at the top left. |
 |
Fig. (3) A Screen shot of the GUI for visual retrieval of fracture cases. |
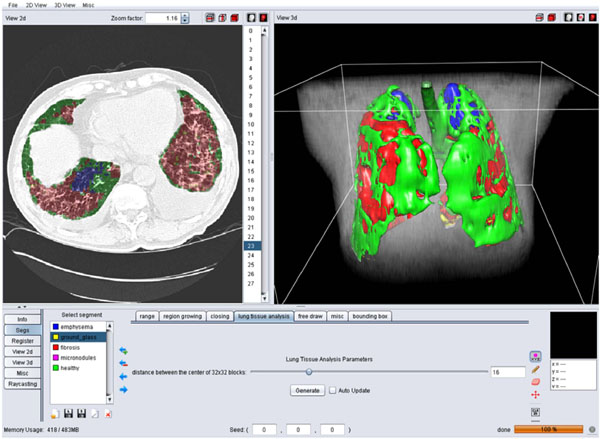 |
Fig. (4) A screen shot of the GUI for the 3D categorization of the lung tissue. |
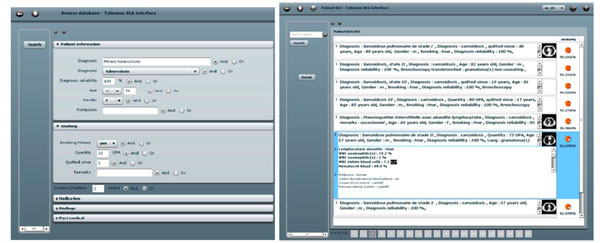 |
Fig. (5) Left: the query interface for clinical parameters. Right: a ranked list of retrieved cases. |
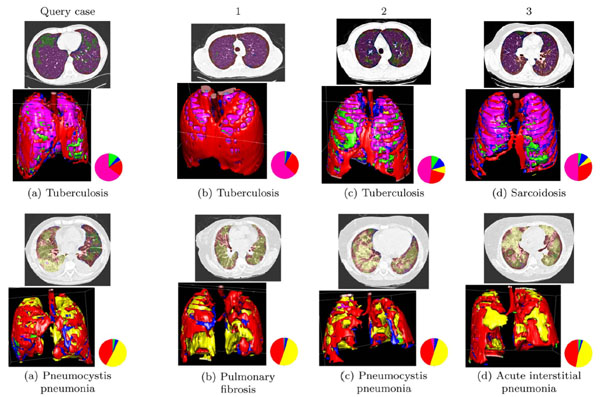 |
Fig. (6) Two examples of a 3D query and retrieved results. |
- Center localization: At first, the centers of the epiphyses are localized. This can be attempted automatically, but a manual localization was proven to be more reliable.
- Region extraction: A bounding box is oriented automatically around these centers, scaled and extracted, yielding the query epiphysial ROIs (eROIs) for the CBIR part of the IRMA-BAA.
- Case comparison: With each extracted eROI, a query by example is send to the case database, which contains complete radiographs, corresponding eROIs, and meta-information such as gender, ethnic origin, chronological age, and the validated bone age from expert readings. For each query, the K most similar eROIs are returned with similarity scoring and validated bone age. Currently, the similarity is determined by cross-correlation, the image distortion model, and the Tamura texture features [41Fischer B, Welter P, Grouls C, Guenther RW, Deserno TM. Bone age assessment by content-based image retrieval and case-based reasoning Proc SPIE 2011; 7963].
- Age assessment: The overall bone age is predicted from the similarities of the K retrieved eROIs and the validated bone ages:
Here, R is the number of eROIs used for the query, agevalidated(r,k) is the validated bone age of the k-th similar
eROI to the r-th query eROI, while ∂ (r, k) provides the corresponding similarity. In (Eq.1) only corresponding bones are compared, e.g., an eROI of distal phalanges of the index finger is compared solely to other eROIs of distal phalanges of the index finger.
Since the IRMA system already provides mechanisms for the inclusion of CBIR into web-based interfaces [42Deserno TM, Güld MO, Plodowski B, et al. Extended query refinement for medical image retrieval J Digit Imaging 2008; 21(3): 280-89.], the BAA-application makes use of these features and a research prototype is available as an online demo2. The prototype allows BAA for a radiograph of the demo database or the analysis of a user-uploaded image. The integration into clinical information systems can be achieved by Digital Imaging and Communications in Medicine (DICOM) Hosted Applications and DICOM Structured Reporting.
The result of the CBIR-based age estimation is presented to the user (Fig. 2 ). Query image and extracted eROIs are shown at the top-most area of the web-based interface. Their most similar counterparts retrieved from the database are shown below (scrollable) in decreasing similarity and with the validated bone age. The estimated bone age is shown below the query image. If the query image is contained in the demo database, the validated bone age is also provided. A click on one of the thumbnails opens the full resolution image. The display mode can also be switched to show the hands belonging to the retrieved eROIs.
). Query image and extracted eROIs are shown at the top-most area of the web-based interface. Their most similar counterparts retrieved from the database are shown below (scrollable) in decreasing similarity and with the validated bone age. The estimated bone age is shown below the query image. If the query image is contained in the demo database, the validated bone age is also provided. A click on one of the thumbnails opens the full resolution image. The display mode can also be switched to show the hands belonging to the retrieved eROIs.
Validation
In order to estimate the potential for clinical use, the research prototype was evaluated in terms of the mean absolute prediction error of the estimated bone age in comparison to the validated bone age. As ground truth, the publicly available hand atlas provided by the University of Southern California (USC) is used, providing 1,102 radiographs with gender, ethnic origin (Caucasian, African American, Asian, Hispanic), and bone age readings by two experienced radiologists. The mean of the two readings is defined as the validated bone age in (Eq. 1). In leaving-one-out experiments, a mean absolute error of 0.97 years and a variance of 0.63 are observed over all ages and regardless of gender.
Summary and Perspective
Although the CBIR-CAD for bone age assessment performs less exact than the commercial application BoneXpert, where a root mean square error of 0.61 years is reported [40Thodberg HH, Kreiborg S, Juul A, Pedersen KD. The bone xpert method for automated determination of skeletal maturity IEEE Trans Med Imaging 2009; 28(1): 52-66.], it provides the radiologist not only the estimated age as a number but also the images to compare with. Further developments in the similarity computation, the inclusion of carpal bones and a gender-specific retrieval are expected to improve the performance. From a CBIR perspective, relevance feedback on the results should be established allowing the radiologist to express his level of agreement to the retrieved images, and restart the CBIR cycle (query refinement) to improve the age estimation.
3.1.2. Bone Fractures
The application described in this section is a case-based CBIR system for surgical planning of bone fractures developed at the HUG [43Zhou X, Stern R, Müller H. Multiscale salient point-based retrieval of fracture cases Proc SPIE 2011; 7967]. A live demonstration of this system was presented at the CADdemo@CARS 2010 in Geneva.
Background and Objective
Fractures are common injuries, and some more complicated fractures require a surgical intervention. Statistics from the Swiss Federal Office of Statistics (SFOS) show that in Switzerland in 2000, 62,535 hospitalizations were due to fractures, and the direct medical cost of hospitalization of patients with osteoporosis and/or related fractures was 357 million of Swiss Francs. Helping the surgeons to plan an intervention in an optimal manner is important from both clinical and economic aspects. Images play an important role in the decision making and the judgment of a fracture. The most common imaging technique used is X-ray, but CT, MRI, and 3-dimensional reconstructions can be required to precisely identify and understand complex configurations. The choice of the surgical technique (e.g., plate, screws) to apply is based on standardized fracture classifications provided by the Orthopaedic Trauma Association (OTA), as well as the personal experience of the surgeon, the latter being often limited to a subgroup. Therefore, it can be beneficial to have access to similar past cases including follow ups to compare, which method might be the most adapted one in a particular situation. However, searching for similar cases in large image databases using keywords is not optimal as it requires prior manual annotation of all images, which is time-consuming and error-prone. Content-based visual information retrieval is used in this application to allow surgeons to submit images of a case to be operated as query and find similar cases ordered by visual similarity.
Methods and Application
The study is based on an image database built at the surgery department of the HUG containing 2,693 fracture cases associated with 43 different fracture types based on OTA classification. Beside images, a few clinical attributes such as age, sex, implant type and exact diagnosis are available in eXtensible Markup Language (XML) files. The fracture retrieval engine is based purely on visual information extracted from image content and the diagnosis information is used only for evaluation. Images are indexed using a bag of visual features strategy, where 1600 local descriptors based on scale-invariant feature transform (SIFT) [44Lowe DG. Object recognition from local scale-invariant features In: Proc Int Conf Computer Vision. Greece: Kerkyra 1999; pp. 1150-7.] are obtained at fixed positions (40x40 grid). Descriptors located at the corresponding position are gathered for variance analysis. Positions are ranked based on the variance as low variance leads to low saliency of the associated position. Only descriptors associated to the best 500 positions were taken. K-means clustering is used to reduce the feature space, which led to 1000 clusters. Consequently, each image is represented by a histogram of 1000 features.
To execute queries, a case-based query interface was developed (Fig. 3 ). Similarity measurement based on histogram intersection is used to rank the returned cases. Both query and results are case-based and contain multiple images. A fusion strategy based on a mix of sum and max operators is in use.
). Similarity measurement based on histogram intersection is used to rank the returned cases. Both query and results are case-based and contain multiple images. A fusion strategy based on a mix of sum and max operators is in use.
Validation
To evaluate the retrieval performance, a maximum of ten cases per fracture type in the database (i.e., class) were used for queries. Each query used all images of one case. Only cases of the same fracture type were considered as relevant.
Average retrieval precisions of the system using the selected subset of queries are of 0.73, 0.29 and 0.21 for P@1, P@10 and P@30, respectively. Although performance is strongly dependent to the number of existing cases per diagnosis, one observed challenge is to distinct two types of factures in the same bone. Typically the precisions for distal fractures are often worse due to the existence of a large number of diaphysis fractures and proximal fractures in the same bone.
Summary and Perspective
A case-based fracture retrieval engine is available online as a treatment planning tool for the surgeons at the HUG. The main novelty of the system is to allow for case-based queries using multiple images. An evaluation of the retrieval precisions based on diagnosis information was performed. The results showed that the system can retrieve fracture cases from the correct bone, but has limited performance for characterizing the exact location and type of fracture (e.g., partial, complex). Involving the user in the loop to refine returned results and using ROIs for queries may help to improve user satisfaction.
3.1.3. Interstitial Lung Diseases
A diagnosis aid framework for HRCT images from patients with ILDs developed by the HUG was presented as a live demonstration at the CADdemo@CARS 2010 in Geneva [45Depeursinge A, Vargas A, Gaillard F, et al. Case-based lung image categorization and retrieval for interstitial lung diseases: clinical workflows Int J Comput Assist Radiol Surg 2011. in press].
Background and Objective
The interpretation of HRCT of the chest from patients with ILD is often challenging with numerous differential diagnoses [46Webb WR, Müller NL, Naidich DP. High-resolution CT of the Lung. Philadelphia: Lippincott Williams & Wilkins 2001.]. Whereas automatic detection and quantification of the lung tissue patterns showed promising results in several studies, its aid for the clinicians is limited to the challenge of image interpretation, letting the radiologists with the problem of the final histological diagnosis. In this application, a hybrid CBIR-CADx was developed. In a first step, a 3D map of the lung tissue resulting from texture classification reduces the risk of omission and ensures the reproducibility of the diagnosis by drawing the radiologists’ attention on diagnostically interesting events. Then, content-based retrieval of similar cases based on the 3D map of lung tissue provides comprehensive information concerning histological diagnoses in the form of HRCT image series with annotated regions in 3D and contextual clinical parameters.
Methods and Application
So far, 85 cases associated with 13 frequent diagnoses of ILDs that underwent an HRCT examination were retrospectively collected at the HUG. Based on each histological diagnosis, the most discriminative clinical parameters for the establishment of the differential diagnostic were kept, resulting in 159 clinical attributes. For each case having a biopsy-based or equivalent (e.g. tuberculin skin test for tuberculosis, Kveim test for sarcoidosis, ...) proven diagnosis, the HRCT image series were simultaneously annotated by two radiologists with more than 15 years of experience. The 159 clinical attributes were filled based on their availability in the electronic health record within a time interval of two weeks around the date of the HRCT image series.
Block-wise texture analysis based on tailored wavelet transforms and support vector machines is used for the categorization of the lung tissue in HRCT. The considered classes of lung tissue are healthy, emphysema, ground glass, fibrosis and micronodules and were selected as being the most represented lung tissue. The segmentation results for the five classes are color-coded to constitute a three-dimensional map of the lung tissue sorts as diagnostic aid (Fig. 4 ).
).
Based on the proportions of the five lung tissue types as well as clinical parameters, a multimodal similarity measure is introduced to enable case-based retrieval. The multimodal inter-case measure used is a linear combination of Euclidean distances that combines clinical parameters of two importance levels as well as the respective volumes of lung tissue sorts output from the texture classification. The interfaces for query formulation and result visualization provide pictograms of the images (Fig. 5 ). The retrieval is annotated with a pie chart visualizing the relative occurrence of different tissue types (Fig. 6
). The retrieval is annotated with a pie chart visualizing the relative occurrence of different tissue types (Fig. 6 ).
).
Validation
In order to evaluate the performance of both texture analysis framework for classifying the lung tissue regions and case-based retrieval, a leave-one-patient-out cross-validation (LOPO CV) of 85 image series was performed. Global geometric and arithmetic mean accuracies of 88.5% and 71.6% are achieved respectively for the lung tissue categorization. Confusions between healthy and micronodules patterns are observed as some of the bronchovascular structures are mixed with small nodules and inversely. The case-based retrieval precision was evaluated based on the diagnosis of the retrieved cases using the seven most represented histological diagnoses. Mean retrieval precisions of 71.05, 47.22 and 39.71 at ranks 1, 5 and 10 respectively are obtained with a LOPO CV.
Summary and Perspective
The main novelty of the system is to allow for submitting 3D queries with contextual clinical parameters. It also shows how CADx and CBIR can be complementary both on the algorithmic and on the user side. Limitations occur as low retrieval performance is obtained when based on visual similarity only, also because the link between visual similarity of two HRCT scans and their associated diagnoses is not straightforward (Fig. 6 ). Another limitation of the system lies in the computing time necessary for creating the 3D map of lung tissue, which is of approximately 20 minutes. High-performance computing may help reducing the computational time of this online task.
). Another limitation of the system lies in the computing time necessary for creating the 3D map of lung tissue, which is of approximately 20 minutes. High-performance computing may help reducing the computational time of this online task.
3.1.4. Mammographic Mass Retrieval
A mammographic mass retrieval platform developed by the Georgetown University Medical Center and University of Michigan Medical Center was presented in SPIE CAD workshops in 2009 and 2010 [36Tao Y, Lo SC, Hadjiski L, Chan HP, Freedman MT. BI-RADS guided mammographic mass retrieval Proc SPIE 2011; 7963: 2H.].
Background and Objective
Breast cancer is among most important causes of mortality of women. Imaging examinations allow early detection of cancerous masses, which can be consequently removed to avoid cancer generalization and metastases. The most common imaging technique used is digital mammography that enables highly detailed visual assessment of breast tissue with low radiation dose delivery. However, only 13-29% of suspicious masses are malignant [47Yankaskas BC, Knelson MH, Abernethy ML, Cuttino JT Jr, Clark RL. Needle localization biopsy of occult lesions of the breast: experience in 199 cases Invest Radiol 1988; 23(10): 729-33.] and a high level of experience is required for accurate staging that is based on the visual appearance of the mass (e.g., shape regularity, texture). To ensure reproducibility of the image interpretation, standardized reporting rules are available, such as the Breast Imaging-Reporting and Data System (BI-RADS) [48Orel SG, Kay N, Reynolds C, Sullivan DC. BI-RADS categorization as a predictor of malignancy Radiology 1999; 211(3): 845-50.].
In clinical practice, experienced radiologists often refer to mental personal images of past cases to evaluate the malignancy of the observed masses, which is subject to inter- and intra- observer variations depending on level of experience and eyestrain. Therefore, using CBIR to systematically provide similar masses with proven diagnoses can assist less experienced radiologists and refresh mental memories of the experienced ones.
Methods and Application
In a first step, the system detects mass candidates using a pixelwise analysis of the mammogram [49Tao Y, Lo SC, Freedman MT, Makariou E, Xuan J. Multilevel learning-based segmentation of ill-defined and spiculated masses in mammograms Med Phys 2010; 37(11): 5993-6002.]. The shape of each detected mass is characterized based on its likelihood to belong to the following BI-RADS-related categories: regular, irregular and lobulated. To allow this characterization, a classifier outputs likelihood of each category based on features describing the boundary-based shape. The latter consists of: third order moments, curvature scale space descriptors (CCSD), radial length statistics, and region-based shape features including compactness, solidity, and eccentricity. Another four BI-RADS-related categories (i.e., circumscribed, microlobulated, indistinct and spiculated) are used to characterize the masses and are predicted using texture features derived from gray level co-occurrence matrices, intensity-based statistics and acutance histograms. The similarity between two masses is computed using locally linear embedding [50Roweis ST, Saul LK. Nonlinear dimensionality reduction by locally linear embedding Science 2000; 290(5500): 2323-6.] in the hyperspace of all seven BI-RADS-related categories.
The system provides the user with the necessary functionality to query using radiologist annotated keyword, specific image features, or using both of them at the same time (Fig. 7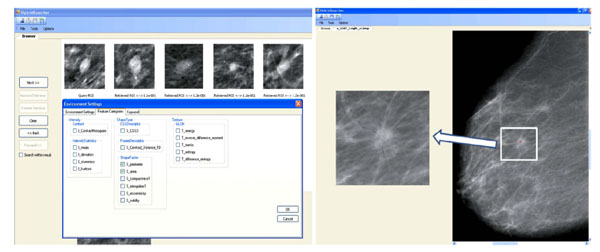 ). The system also allows the user to select the image features used to characterize masses.
). The system also allows the user to select the image features used to characterize masses.
Validation
The system is validated on two datasets. The first one consisted of 415 mammographic biopsy-proven masses, in which 244 were malignant and 171 were benign. A receiver operating curve (ROC) analysis showed that the retrieval performance had an area under the ROC curve of 0.75±0.03 for the five first retrieved masses. The second dataset consisted of 476 masses (219 malignant and 257 benign). 10 random test/train partitions are used to evaluate the retrieval performance where the test set consisted of 76 masses (38 malignant and 38 benign), and the training set of the 400 remaining. An area under the ROC curve of 0.80±0.06 was obtained for the five first retrieved masses.
Summary and Perspective
The development of the system is strongly focused on image features able to catch the subtle differences between benign and malignant with performance close to experienced readers. A potential limitation of the system lies in the fact that radiologists may not be able to understand the various influences of advanced image features (e.g., Fourier descriptors, gray-level co-occurrence matrices) on the retrieval performance.
3.2. System Comparison
A direct comparison of the four systems would not lead to meaningful results, as the contributions in medical CBIR are driven by the various application needs. For instance, only the application for interstitial lung diseases would benefit from possibility to submit three-dimensional queries. Moreover, a comparison of the retrieval performance would be of little interest, since the latter strongly depends on the difficulty of the medical task as well as the database and methodology used for validation. However, the systems can be compared in terms of their “user readiness”, as defined in Section 2.13. The strengths and possible improvements of the systems relatively to the addressed medical application are summarized in Table 4.
The IRMA system for bone age assessment provides contributions in the GUI and clinical workflows. The visualization of the results enables transparency of the methods by displaying the epiphyseal regions used for retrieval and age prediction. The processing workflows correspond to the human approach for BAA, which improves the system’s intuitiveness. Several possible developments could still improve the user readiness of the system. In particular displaying intermediate steps allowing for selecting the epiphyseal regions used for retrieval. Using clinical parameters (e.g., gender, ethnics) as well as implementing query refinement and relevance feedback could also be beneficial to enable quicker convergence to sought cases.
The fracture case application is the only one enabling case-based multi-image queries. This important feature allows including the intra-class variations directly in the query and subsequently improves the level of semantics expressed by the user. The system suffers from the complexity of the medical task, since finding the fracture region is challenging. Therefore, this system would highly beneficiate from ROI-based queries, where the user could use ROIs from several images to best represent the fracture class. In addition, including clinical parameters such as the patient’s age in the query as well as enabling user feedback would be beneficial to improve user readiness of the system.
The clinical requirements of the application for interstitial lung diseases are challenging. The three-dimensionality of HRCT images used and the non-triviality of the links between visual appearance of the lung tissue and histological diagnoses of ILDs open various challenges for developing a CBIR system fulfilling user needs. Some issues are addressed, as the system allows submitting 3D queries and enables system transparency by displaying intermediate image analysis results in the form of a 3D map of categorized lung tissue. It also permits to formulate multimodal queries with a large set of clinical parameters related to ILDs. Additional work is required to develop visual features characterizing three-dimensional lung textures in isotropic multi-detector row computed tomography (MDCT) image series to closely express human perception of lung textures and exploit the three-dimensionality of the original data. Implementing multiple ROI-based queries and user relevance feedback functionalities are also important issues to reduce the semantic gap. High performance computing is required to reduce online computational time required to create the 3D map of categorized lung tissue.
The mammographic mass retrieval system investigated several developments in terms of visual features in order to characterize the subtle differences among benign and malignant breast masses. It allows submitting mixed text and visual queries based on automatically detected ROIs. Several improvements are possible in clinical workflows to enhance user interaction. For instance involving the user in the selection of the candidate masses, enabling for submitting multiple examples of the same mass (over time for example), allowing several iterations with user relevance feedback could enhance the rapidity of convergence to sought masses. The understandability of the system is not trivial for a radiologist user, especially for selecting visual features to be used for retrieval, since clinicians may not be familiar with the impact of Fourier descriptors or gray-level co-occurrence matrices on the quality of retrieval.
3.3. . Major Gap Identification
Although several strengths have been emphasized for the four prototype systems included in this analysis, they still are prototypes with a certain degree of readiness for clinical use. Table 5 reflects the systems with respect to the gaps [20Deserno TM, Antani S, Long R. Ontology of gaps in content-based image retrieval J Digit Imaging 2009; 22(2): 202-15.].
In general, the content gaps including the semantic gap are bypassed via the specificity of the systems and including some manual components in locating the region of interest. While global 2D features in 3D data have been used for a long time, the feature gaps obviously have been closed by all of the systems. Only one system is a 3D application, and here a multi-scale implementation might speed up the process and simplify the user interaction. So far, the group of performance gaps has been addressed only partly. All systems provide an application that has been evaluated quantitatively on a comprehensive set of ground truth data, guaranteeing its effectiveness. On the other hand, however, integration and indexing gaps have not been dealt sufficiently yet. The same holds for the systems’ usability. Man machine interaction for query formulation and refinement as well as relevance feedback may need improvement to successfully migrate the prototypes into clinical practice.
4. DISCUSSIONS AND CONCLUSIONS
In this paper, we have systematically searched for CBIR applications in CAD with high level of “user readiness”. In total, 70 system presentations on internationally recognized workshops and conferences were analyzed and four CBIR prototypes were identified. The applications are in the fields of bone age assessment, bone fracture, interstitial lung diseases, and screening mammography. All of these fields were identified as having a key role for medical image processing and CAD, including a long history of developing reference databases [51Horsch A, Prinz M, Schneider S, Sipila O, et al. Establishing an international reference image database for research and development in medical image processing Method Inform Med 2004; 43(4): 409-12.]. Accordingly, optical imaging (microscopy and endoscopy) and medical ultrasound imaging in colon and esophagus might be the future fields for CBIR prototype development.
With respect to the gaps [20Deserno TM, Antani S, Long R. Ontology of gaps in content-based image retrieval J Digit Imaging 2009; 22(2): 202-15.], all systems – based on the training data –annotate query images with meaningful terms such as age, tissue type, or diagnosis, and by this, adequately bridge the semantic gap. However, the use context is very specific, not allowing the transfer of the system into other domains in clinical practice. Future CBIR-CAD shall aim at more generic architectures, principles and interfaces to support rapid prototyping for other applications.
Nonetheless, all of the prototypes have bridged the various feature gaps, and efficiency was proven on relatively large reference databases and ground truth images. This can be seen as a prerequisite for any prototype in the field of CBIR-based CAD. However, all of the prototypes still need improvement in user interaction and system integration. So far, stand-alone systems are demonstrated that are not appropriately integrated in the hospital information system environments. To become part of the medical information system environment that is usually operated in clinical practice, seamless integration is required on different levels [52Leisch E, Sartzetakis S, Tsiknakis M, Orphanoudakis SC. A framework for the integration of distributed autonomous healthcare information systems Med Inform 1997; 22(4): 325-35.,53Winter A, Haux R, Ammenwerth E, Brigl B, Hellrung N, Jahn F. Health Information Systems: Architectures and Strategies. Berlin: Springer 2010.]:
- data integration is achieved if any data stored in the prototype must not be entered more than once;
- functional integration is obtained if any services provided by the prototype can be used from any other module of the information system requiring this particular service;
- presentation integration is obtained if data presented in the prototype and other modules of the entire system appears in a unified way and style; and
- context integration is present if specific settings such as the selection of a certain patient or image, which is done in one module, is passed automatically to the prototype when it is called, and vice versa.
Furthermore, the results obtained from the prototype must be stored in the picture archiving and communication system (PACS). DICOM Hosted Application and DICOM Structured Reporting might be suitable concepts in bridging the integration gap [54Welter P, Riesmeier J, Fischer B, et al. Bridging the integration gap from imaging to information systems J Am Med Inform Assoc 2011. in press].
In conclusion, the usability of the prototypes bears several options for improvement. Whereas some of the improvements may not be applicable to disease- or organ-specific CBIR systems, most of the strengths of the prototypes analyzed in this work are applicable to any medical application where CBIR is relevant. Intuitiveness of a system is an unconditional prerequisite and should not rely on user’s computer literacy level. The workflows of the system must be as close as possible to common clinical practices. Query formulation is also an important issue to allow clinicians to adequately express their search intentions. Among the four systems, ROI-based queries, multiple image/ROI queries as well as mixed text/visual queries showed to allow efficient query formulation. ROI-based queries allow selecting visual patterns corresponding to the targeted classes with high precision whereas multiple image/ROI queries express intra-class variations. Extended query refinement and relevance feedback are concepts that need integration in the prototypes [42Deserno TM, Güld MO, Plodowski B, et al. Extended query refinement for medical image retrieval J Digit Imaging 2008; 21(3): 280-89.]. When applicable, showing intermediate image analysis results used for retrieval (e.g., automatically segmented objects or regions) provides transparency of the methods. At last, rapid online response of the system is also an important factor for a successful adoption in the stressful environments that hospitals are.
NOTES
CONFLICT OF INTERESTD
None declared.
ACKNOWLEDGEMENTS
This work was supported by the European Union, Marie Curie Actions, International Research Staff Exchange Scheme (FP7-PEOPLE-2010-IRSES, Project No 269124, Warehousing images in the digital hospital (WIDTH): interpretation, infrastructure, and integration). Partial support for this work was obtained from the Swis National science foundation (MANY project) and the European Union (Khresmoi project).