- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Deep Sequencing Details the Cross-over Map of Chimeric Genes in Two Porcine Reproductive and Respiratory Syndrome Virus Infectious Clones
Nanhua Chen1, 2, *, Ranjni J. Chand2, Raymond R. R. Rowland2
Abstract
Background:
Recombination is an important contributor to the genetic diversity of most viruses. A reverse genetics system using green fluorescence protein (GFP)- and enhanced GFP (EGFP)-expressing infectious clones was developed to study the requirements for recombination. However, it is still unclear what types of cross-over events occurred to produce the viable offspring.
Objective:
We utilized 454 sequencing to infer recombination events in this system.
Method:
Two porcine reproductive and respiratory syndrome virus (PRRSV) infectious clones, P129-EGFP-97C and P129-GFPm-d (2-6), were co-transfected into HEK-293T cells. P129-EGFP-97C is a fully functional virus that contains a non-fluorescent EGFP. P129-GFPm-d (2-6) is a defective virus but contains a fluorescent GFPm. Successful recombination was evident by the appearance of fully functional progeny virus that expresses fluorescence. Total RNA was extracted from infected cells expressing fluorescence, and the entire fluorescent gene was amplified to prepare an amplicon library for 454 sequencing.
Results:
Deep sequencing showed that the nucleotide identities changed from ~37% (in the variable region from 21nt to 165nt) to 20% (T289C) to ~38% (456-651nt) then to 100% (672-696nt) when compared to EGFP. The results indicated that cross-over events occurred in three conserved regions (166-288nt, 290-455nt, 652-671nt), which were also supported by sequence alignments. Remarkably, the short conserved region (652-671nt) showed to be a cross-over hotspot. In addition, four cross-over patterns (two single and two double cross-over) might be used to produce viable recombinants.
Conclusion:
The reverse genetics system incorporating the use of high throughput sequencing creates a genetic platform to study the generation of viable recombinant viruses.
Article Information
Identifiers and Pagination:
Year: 2017Volume: 11
Issue: Suppl-1, M2
First Page: 49
Last Page: 58
Publisher Id: TOVJ-11-49
DOI: 10.2174/1874357901711010049
Article History:
Received Date: 06/10/2016Revision Received Date: 01/11/2016
Acceptance Date: 06/02/2017
Electronic publication date: 30/06/2017
Collection year: 2017
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: https://creativecommons.org/licenses/by/4.0/legalcode. This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
* Address correspondence to this author at the College of Veterinary Medicine, Yangzhou University, Yangzhou, Jiangsu 225009, P.R. China, Tel: +86-514-87988323; Fax: +086-514-87972218; E-mail: chnhlh@126.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 06-10-2016 |
Original Manuscript | Deep Sequencing Details the Cross-over Map of Chimeric Genes in Two Porcine Reproductive and Respiratory Syndrome Virus Infectious Clones | |
INTRODUCTION
Recombination occurs in most RNA viruses and has a major impact on their diversification and evolution. More significantly, recombination has also been associated with the emergence of new viruses, increases in virulence and pathogenesis, and the evasion of host immune responses [1Perez-Losada M, Arenas M, Galan JC, et al. Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infection, genetics and evolution journal of molecular epidemiology and evolutionary genetics in infectious diseases 30C 2015; 296-307., 2Simon-Loriere E, Holmes EC. Why do RNA viruses recombine? Nat Rev Microbiol 2011; 9: 617-26.]. Porcine reproductive and respiratory syndrome virus (PRRSV) is the most costly swine virus worldwide, which is an enveloped, positive-sense, single-stranded RNA virus [3Lunney JK, Fang Y, Ladinig A, et al. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): Pathogenesis and Interaction with the Immune System. Annu Rev Anim Biosci 2016; 4: 129-54.]. Recombination in RNA viruses is a process to form chimeric offspring from parental genomes of mixed origin, which requires co-infection or super-infection of a cell with at least two viruses. RNA recombination is based on RNA-dependent RNA polymerase (RdRp) template switches that may occur during either genomic RNA replication or subgenomic mRNA synthesis [4Pasternak AO, Spaan WJ, Snijder EJ. Regulation of relative abundance of arterivirus subgenomic mRNAs. J Virol 2004; 78: 8102-13.]. The factors that influence template switching include RNA secondary structures and sequence similarity between donor and acceptor templates [5Galetto R, Giacomoni V, Veron M, Negroni M. Dissection of a circumscribed recombination hot spot in HIV-1 after a single infectious cycle. J Biol Chem 2006; 281: 2711-20., 6Zhang J, Temin HM. Retrovirus recombination depends on the length of sequence identity and is not error prone. J Virol 1994; 68: 2409-14.].
Recombination is a common phenomenon between PRRSV isolates in the field [7Martin-Valls GE, Kvisgaard LK, Tello M, et al. Analysis of ORF5 and full-length genome sequences of porcine reproductive and respiratory syndrome virus isolates of genotypes 1 and 2 retrieved worldwide provides evidence that recombination is a common phenomenon and may produce mosaic isolates. J Virol 2014; 88: 3170-81.]. The occurrence of PRRSV recombination was first suggested by the phylogenetic analysis of field isolates [8Kapur V, Elam MR, Pawlovich TM, Murtaugh MP. Genetic variation in porcine reproductive and respiratory syndrome virus isolates in the midwestern United States. J Gen Virol 1996; 77(Pt 6): 1271-6.]. Among PRRSV isolates, intra-genotype recombination is frequent but no inter-genotype recombination has been reported [9Chen N, Yu X, Wang L, et al. Two natural recombinant highly pathogenic porcine reproductive and respiratory syndrome viruses with different pathogenicities. Virus Genes 2013; 46: 473-8.-16Lu WH, Tun HM, Sun BL, et al. Re-emerging of porcine respiratory and reproductive syndrome virus (lineage 3) and increased pathogenicity after genomic recombination with vaccine variant. Vet Microbiol 2015; 175: 332-40.]. PRRSV could undergo homologous recombination with the frequency from <2% up to 10% in vitro and ~38% (133/352) in vivo [12Yuan S, Nelsen CJ, Murtaugh MP, Schmitt BJ, Faaberg KS. Recombination between North American strains of porcine reproductive and respiratory syndrome virus. Virus Res 1999; 61: 87-98., 14Liu D, Zhou R, Zhang J, et al. Recombination analyses between two strains of porcine reproductive and respiratory syndrome virus in vivo. Virus Res 2011; 155: 473-86.]. The most widely accepted model of PRRSV recombination is copy-choice model [2Simon-Loriere E, Holmes EC. Why do RNA viruses recombine? Nat Rev Microbiol 2011; 9: 617-26., 17Lai MM. RNA recombination in animal and plant viruses. Microbiol Rev 1992; 56: 61-79.].
Conventional methods to detect PRRSV recombination are based on PCR and sequencing followed by data analysis using recombination detection programs, such as RDP and SimPlot [18Lole KS, Bollinger RC, Paranjape RS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 1999; 73: 152-60., 19Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: A flexible and fast computer program for analyzing recombination. Bioinformatics 2010; 26: 2462-3.]. A major limitation of these PCR based methods is that all genomic, subgenomic and defective RNAs could serve as templates for PCR amplification. Therefore, the viability of the recombinants identified by them is unknown. To address this limitation, we developed a new in vitro system targeting at recombination events that present in viable offspring [20Chand R. Study of Recombination in PRRSV using a novel in-vitro system. 2013. PhD Thesis. Kansas State University, Manhattan, KS.]. This reverse genetics system uses green fluorescence protein (GFP)- and enhanced GFP (EGFP)-expressing PRRSV infectious clones to study recombination in the chimeric genes that are nonessential for virus replication. Successful recombination is evidenced by generating a viable fluorescent virus from the co-transfection of a non-fluorescent viable virus with a mutation in EGFP (P129-EGFP-97C) and a fluorescent defective virus (P129-GFPm-d (2-6)) (Fig. 1 ). However, what types of cross-over events occurred to produce viable viruses in this system are not clarified yet. Here we took advantage of high throughput sequencing to assess the locations of all cross-over events between EGFP and GFPm genes and explore the cross-over patterns that are potentially utilized to produce viable recombined viruses.
). However, what types of cross-over events occurred to produce viable viruses in this system are not clarified yet. Here we took advantage of high throughput sequencing to assess the locations of all cross-over events between EGFP and GFPm genes and explore the cross-over patterns that are potentially utilized to produce viable recombined viruses.
MATERIALS AND METHOD
Sample Preparation
HEK-293T cells were propagated and maintained in Minimum Essential Medium Eagle (1×MEM) (Fisher Scientific) with 7% Fetal Bovine Serum (FBS) (Gibco), 80 U/ml Penicillin-Streptomycin (Gibco) and 0.3 μg/ml Fungizone Antimycotic (Gibco) at 37°C with 5% CO2 [21Guo R, Katz BB, Tomich JM, Gallagher T, Fang Y. Porcine reproductive and respiratory syndrome virus utilizes nanotubes for intercellular spread. J Virol 2016.]. About 80% confluent HEK-293T cells were co-transfected with two PRRSV infectious clones: P129-EGFP-97C and P129-GFPm-d (2-6), using Fugene HD transfection reagent (Promega) according to the recommended protocol. These two infectious clones are derived from the DNA-launched P129-GFP infectious clone [22Pei Y, Hodgins DC, Wu J, et al. Porcine reproductive and respiratory syndrome virus as a vector: Immunogenicity of green fluorescent protein and porcine circovirus type 2 capsid expressed from dedicated subgenomic RNAs. Virology 2009; 389: 91-9., 23Welch SK, Jolie R, Pearce DS, et al. Construction and evaluation of genetically engineered replication-defective porcine reproductive and respiratory syndrome virus vaccine candidates. Vet Immunol Immunopathol 2004; 102: 277-90.] but with distinct characteristics. P129-EGFP-97C is a fully functional P129 virus with a non-fluorescent EGFP gene, the result of a C289T nucleotide substitution in the fluorophore active site of EGFP [24Dinh PX, Panda D, Das PB, Das SC, Das A, Pattnaik AK. A single amino acid change resulting in loss of fluorescence of eGFP in a viral fusion protein confers fitness and growth advantage to the recombinant vesicular stomatitis virus. Virology 2012; 432: 460-9.]. P129-GFPm-d (2-6) is a defective virus that lacks ORFs 2-6, but contains a fluorescent GFPm gene. GFPm is a chimeric gene that contains a middle EGFP sequence (290bp) flanked on each side by sequence derived from GFP (EGFP and GFP share only 83% nucleotide identity). Fully functional viruses that express fluorescence can be generated by recombination between these two infectious clones in fluorescent genes. Therefore, after 48 hours of co-transfection, the supernatant was used to infect 100% confluent Marc-145 cells and green fluorescent plaques could be observed at 72 hours post infection (hpi). Due to the low frequency of recombination (~0.3%), enrichment was performed using fluorescence-activated cell sorting (FACS) for cells that express green fluorescence with the MoFlo XDP Cell Sorter (Beckman Coulter). After two rounds of enrichment, roughly 80% of cells carrying the fluorescent viruses were obtained [20Chand R. Study of Recombination in PRRSV using a novel in-vitro system. 2013. PhD Thesis. Kansas State University, Manhattan, KS.].
Amplicon Library Preparation
Total RNA was extracted from 100 μl of the sorted cell sample using TRIzol® Reagent (Invitrogen) according to the manufacturer’s protocol and eluted in 50 μl RNase-free water. cDNA was generated by reverse transcription using random hexamer primers from the Transcriptor High Fidelity cDNA Synthesis Kit (Roche). Two sets of primer pairs were utilized in two rounds of PCR for the preparation of an amplicon library (Table 1). Three overlapped regions, which were 369bp, 270bp, and 336bp in length, were amplified in the first round of PCR. A same pair of multiplex identifier (MID) primers was used for the three amplicons in the second round of PCR. The amplicon library was created by these three amplicons as we previously described [25Chen N, Trible BR, Kerrigan MA, Tian K, Rowland RR. ORF5 of porcine reproductive and respiratory syndrome virus (PRRSV) is a target of diversifying selection as infection progresses from acute infection to virus rebound. 2016. Infection, genetics and evolution : Journal of molecular epidemiology and evolutionary genetics in infectious diseases 40: 167-75., 26Chen N, Dekkers JC, Ewen CL, Rowland RR. Porcine reproductive and respiratory syndrome virus replication and quasispecies evolution in pigs that lack adaptive immunity. Virus Res 2015; 195: 246-9.].
Pyrosequencing
The amplicon library was sent to emPCR amplification and 454 sequencing as described previously [25Chen N, Trible BR, Kerrigan MA, Tian K, Rowland RR. ORF5 of porcine reproductive and respiratory syndrome virus (PRRSV) is a target of diversifying selection as infection progresses from acute infection to virus rebound. 2016. Infection, genetics and evolution : Journal of molecular epidemiology and evolutionary genetics in infectious diseases 40: 167-75., 26Chen N, Dekkers JC, Ewen CL, Rowland RR. Porcine reproductive and respiratory syndrome virus replication and quasispecies evolution in pigs that lack adaptive immunity. Virus Res 2015; 195: 246-9.]. Briefly, Lib-L emPCR Kit (Roche) was used for emPCR according to the emPCR Amplification Method Manual. GS FLX Titanium Sequencing Kit XLR70 (Roche) was used for 454 sequencing following the protocol. Reads for each sample were sorted according to the MID. Sequence reads were mapped against the GFPm gene with 454 Life Sciences GS Reference Mapper (Version 2.6). Coverage was calculated and variants were called. Variants were filtered based on the coverage, variant frequency, and homopolymer. Only high confidence single nucleotide variants that have the following features were selected: (1) at least 3 non-duplicate reads have the nucleotide substitution; (2) the substitution frequency is greater than 5%; (3) the substitution is not located at homopolymer sites.
RESULTS
Identification of Cross-over Events
The number of reads for three amplicons were around 6300 ~ 15500. All sequences were compared to the GFPm sequence. Mutations identified by high throughput sequencing are shown in (Table 2). The percentages of mutations identical to EGFP in the first variable region (from 21nt to 165nt) were around 31% to 41%, with 37% in average. Based on 15531 reads, only 20% of the sequences had thymine at position 289, while the other 80% were cytosine, which is identical to GFPm. The second variable region (from 456nt to 651nt) is about 29% to 45% (with the average of 38%) identical to EGFP. The third variable region (from 672nt to 696nt) has 100% identity to EGFP. The changes in the percentages of identities between variable regions after each conserved region indicated that cross-over events occurred in three conserved regions: 166-288nt, 290-455nt, and 652-671nt.
As shown in Fig. (2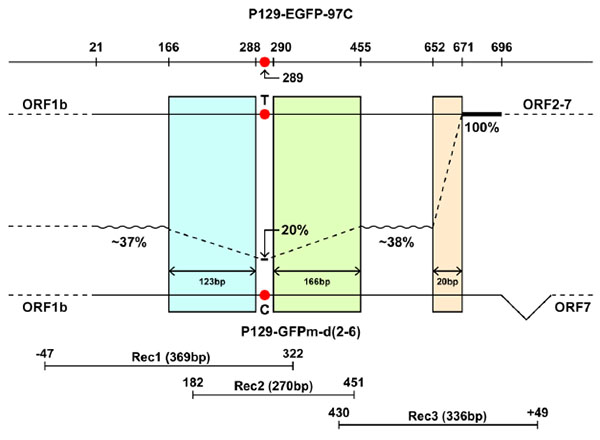 ), the percentage of nucleotide identity decreased from ~37% to 20% in the 123bp-conserved region (166-288nt) when compared to EGFP, which suggested that there was a cross-over occurrence. In addition, the percentage increased from 20% to ~38% in the 166bp-conserved region (290-455nt), suggesting that another cross-over existed in this region. Remarkably, the percentage dramatically changed from 38% to 100%, which meant ~62% of the recombinant viruses proceeded cross-over in the only 20bp-conserved region (652-671nt), suggesting the 20bp-conserved region is a hotspot of cross-over.
), the percentage of nucleotide identity decreased from ~37% to 20% in the 123bp-conserved region (166-288nt) when compared to EGFP, which suggested that there was a cross-over occurrence. In addition, the percentage increased from 20% to ~38% in the 166bp-conserved region (290-455nt), suggesting that another cross-over existed in this region. Remarkably, the percentage dramatically changed from 38% to 100%, which meant ~62% of the recombinant viruses proceeded cross-over in the only 20bp-conserved region (652-671nt), suggesting the 20bp-conserved region is a hotspot of cross-over.
Potential Cross-over Patterns
Four types of cross-over events could occur to produce the recombined, viable, and fluorescent virus. As shown in Fig. (3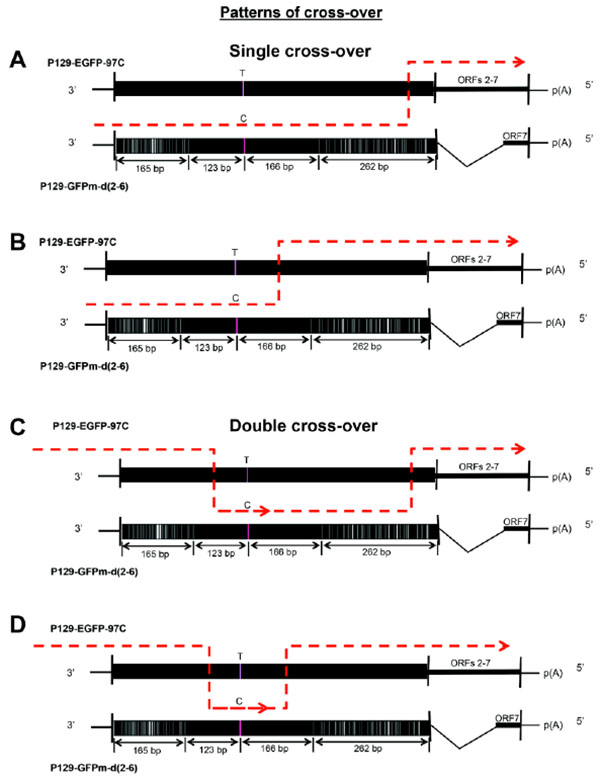 ), there are two types of single recombination events Figs. (3A
), there are two types of single recombination events Figs. (3A and B
and B ), which have cross-over occurring in the 20bp-conserved region (from GFPm to EGFP) and the 166bp-conserved region (from GFPm to EGFP), respectively. Furthermore, there are other two types of double recombination events (Figs. 3C
), which have cross-over occurring in the 20bp-conserved region (from GFPm to EGFP) and the 166bp-conserved region (from GFPm to EGFP), respectively. Furthermore, there are other two types of double recombination events (Figs. 3C and D
and D ). One has the double cross-over occurring in the 123bp-conserved region (from EGFP to GFPm) then in the 20bp-conserved region (from GFPm to EGFP), and another one occurs in the 123bp-conserved region (from EGFP to GFPm) then in the 166bp-conserved region (from GFPm to EGFP).
). One has the double cross-over occurring in the 123bp-conserved region (from EGFP to GFPm) then in the 20bp-conserved region (from GFPm to EGFP), and another one occurs in the 123bp-conserved region (from EGFP to GFPm) then in the 166bp-conserved region (from GFPm to EGFP).
 |
Fig. (3) Four potential cross-over patterns used for producing recombinants. Two single cross-over Figs. (3A  and B and B ) and two double cross-over Figs. (3C ) and two double cross-over Figs. (3C and D and D ) could be used to generate the viable fluorescent recombined viruses in this in vitro reverse genetics system. ) could be used to generate the viable fluorescent recombined viruses in this in vitro reverse genetics system.
|
Although the rate of each recombination pattern could not be identified in this study, sequence alignments provided direct evidence that cross-over events occurred in the 123bp- and 20bp-conserved regions, respectively (Fig. 4 ). The first representative recombinant is identical to GFPm gene before the 123bp-conserved region but becomes identical to EGFP gene from position 289 (Fig. 4A
). The first representative recombinant is identical to GFPm gene before the 123bp-conserved region but becomes identical to EGFP gene from position 289 (Fig. 4A ). And the second representative recombinant is identical to GFPm gene before the 20bp-conserved region but becomes identical to EGFP gene after the conserved region (Fig. 4B
). And the second representative recombinant is identical to GFPm gene before the 20bp-conserved region but becomes identical to EGFP gene after the conserved region (Fig. 4B ). Cross-over events occurring in the 166bp-conserved regions could not be analyzed due to the limit of read length (400bp) of 454-pyrosequencing used in our study.
). Cross-over events occurring in the 166bp-conserved regions could not be analyzed due to the limit of read length (400bp) of 454-pyrosequencing used in our study.
DISCUSSION
By combined utilization of the in vitro reverse genetics system and high throughput sequencing, we inferred recombination events between inserted fluorescent genes in PRRSV infectious clones. Based on thousands of sequences from viable progeny viruses, we found that the nucleotide identities changed between the variable and conserved regions of EGFP/GFPm genes, indicating that the cross-over events occurred in the conserved regions. The advantages of this new method in measuring recombination include: 1) This reporter system targets at detecting viable recombinant viruses. 2) The system tests recombination events in nonessential gene without affecting the virus replication. 3) The co-utilization of in vitro reverse genetics system and deep sequencing reveals all types of cross-over events which occurred in a target gene to produce viable recombinants. Similar reporter systems are generally used to measure the virus recombination [27Levy DN, Aldrovandi GM, Kutsch O, Shaw GM. Dynamics of HIV-1 recombination in its natural target cells. Proc Natl Acad Sci USA 2004; 101: 4204-9., 28Runckel C, Westesson O, Andino R, DeRisi JL. Identification and manipulation of the molecular determinants influencing poliovirus recombination. PLoS Pathog 2013; 9: e1003164.]. The disadvantage of using 454 pyrosequencing in our system is the introduction of errors (error rates ranged from 0.04-0.66%) [29Di Giallonardo F, Zagordi O, Duport Y, et al. Next-generation sequencing of HIV-1 RNA genomes: determination of error rates and minimizing artificial recombination. PLoS One 2013; 8: e74249.], which resulted in the variations of the identities in a same conserved region (Table 2); however, it did not interrupt the evaluation of the obviously changes between conserved regions. In addition, this system was developed to analyze the requirements for generating viable recombinants but not for detecting all recombination events between these two infectious clones, therefore, it could not detect the recombination events in the PRRSV genomes.
The first and second conserved regions (166-288nt and 290-455nt) are only separated by a C289T mutation. And the variation frequencies in two flanked variable regions (21-165nt and 456-651nt) are nearly identical (~37% and ~38%), which arouse suspicion that the lower variation frequency of C289T mutation (20%) might have resulted from mutation or inaccurate sequencing rather than from recombination. However, our results provide strong support that the changes in sequence identity are due to cross-over events. First, the 20% mutation rate of C289T results from 15531 reads (Table 2). Second, the sequencing result of the sample (80% nucleotides at position 289 are cytosine, which is identical to the fluorescent gene GFPm) is consistent with the cell sorting result (about 80% sorted cells express fluorescence). Third, the C289T mutation is highly stable during serial passage in Marc-145 cells [20Chand R. Study of Recombination in PRRSV using a novel in-vitro system. 2013. PhD Thesis. Kansas State University, Manhattan, KS.].
The third conserved region (652-671nt) shows to be a cross-over hotspot and has higher cross-over rate than the above two conserved regions (Fig. 2 ). The result is consistent with Sanger sequencing results that all six individual clones of the whole fluorescent gene from the same co-infection sample had cross-over at the 20bp-conserved region [20Chand R. Study of Recombination in PRRSV using a novel in-vitro system. 2013. PhD Thesis. Kansas State University, Manhattan, KS.]. Although both RNA secondary structure and the length of sequence identity are crucial molecular determinants for recombination [5Galetto R, Giacomoni V, Veron M, Negroni M. Dissection of a circumscribed recombination hot spot in HIV-1 after a single infectious cycle. J Biol Chem 2006; 281: 2711-20., 6Zhang J, Temin HM. Retrovirus recombination depends on the length of sequence identity and is not error prone. J Virol 1994; 68: 2409-14., 28Runckel C, Westesson O, Andino R, DeRisi JL. Identification and manipulation of the molecular determinants influencing poliovirus recombination. PLoS Pathog 2013; 9: e1003164.], RNA secondary structure seems to play a more important role in this case considering that the length of the cross-over hotspot (20bp) is shorter than the other two cross-over regions (123bp and 166bp). In addition, the 20bp-conserved region encoded “117-HMVLLE-222” is located at the junction region of β-sheet strand 11 linking to the loop. Glu222 at the junction point has alternative conformations [30Arpino JA, Rizkallah PJ, Jones DD. Crystal structure of enhanced green fluorescent protein to 1.35 A resolution reveals alternative conformations for Glu222. PLoS One 2012; 7: e47132.]. Previous studies implied that recombination events occurred more frequency at the transcriptional pausing sites or polymerase-binding motifs [5Galetto R, Giacomoni V, Veron M, Negroni M. Dissection of a circumscribed recombination hot spot in HIV-1 after a single infectious cycle. J Biol Chem 2006; 281: 2711-20., 17Lai MM. RNA recombination in animal and plant viruses. Microbiol Rev 1992; 56: 61-79.]. Therefore, the mechanisms responsible for the hotspot of the 20bp-conserved region are probably associated with RNA secondary structure. Notably, recombination events were not detected in the 20bp region when using infectious clones containing EGFP-97C and GFP, sharing 83% nucleotide identity [20Chand R. Study of Recombination in PRRSV using a novel in-vitro system. 2013. PhD Thesis. Kansas State University, Manhattan, KS.]. Recombination events occurred between EGFP-97C and GFPm, which share 91% nucleotide identity, suggesting that the overall nucleotide identity may be also essential to produce recombinants. And another explanation is that the relative positions of the EGFP sequence inserted in GFPm gene may be important [6Zhang J, Temin HM. Retrovirus recombination depends on the length of sequence identity and is not error prone. J Virol 1994; 68: 2409-14.].
). The result is consistent with Sanger sequencing results that all six individual clones of the whole fluorescent gene from the same co-infection sample had cross-over at the 20bp-conserved region [20Chand R. Study of Recombination in PRRSV using a novel in-vitro system. 2013. PhD Thesis. Kansas State University, Manhattan, KS.]. Although both RNA secondary structure and the length of sequence identity are crucial molecular determinants for recombination [5Galetto R, Giacomoni V, Veron M, Negroni M. Dissection of a circumscribed recombination hot spot in HIV-1 after a single infectious cycle. J Biol Chem 2006; 281: 2711-20., 6Zhang J, Temin HM. Retrovirus recombination depends on the length of sequence identity and is not error prone. J Virol 1994; 68: 2409-14., 28Runckel C, Westesson O, Andino R, DeRisi JL. Identification and manipulation of the molecular determinants influencing poliovirus recombination. PLoS Pathog 2013; 9: e1003164.], RNA secondary structure seems to play a more important role in this case considering that the length of the cross-over hotspot (20bp) is shorter than the other two cross-over regions (123bp and 166bp). In addition, the 20bp-conserved region encoded “117-HMVLLE-222” is located at the junction region of β-sheet strand 11 linking to the loop. Glu222 at the junction point has alternative conformations [30Arpino JA, Rizkallah PJ, Jones DD. Crystal structure of enhanced green fluorescent protein to 1.35 A resolution reveals alternative conformations for Glu222. PLoS One 2012; 7: e47132.]. Previous studies implied that recombination events occurred more frequency at the transcriptional pausing sites or polymerase-binding motifs [5Galetto R, Giacomoni V, Veron M, Negroni M. Dissection of a circumscribed recombination hot spot in HIV-1 after a single infectious cycle. J Biol Chem 2006; 281: 2711-20., 17Lai MM. RNA recombination in animal and plant viruses. Microbiol Rev 1992; 56: 61-79.]. Therefore, the mechanisms responsible for the hotspot of the 20bp-conserved region are probably associated with RNA secondary structure. Notably, recombination events were not detected in the 20bp region when using infectious clones containing EGFP-97C and GFP, sharing 83% nucleotide identity [20Chand R. Study of Recombination in PRRSV using a novel in-vitro system. 2013. PhD Thesis. Kansas State University, Manhattan, KS.]. Recombination events occurred between EGFP-97C and GFPm, which share 91% nucleotide identity, suggesting that the overall nucleotide identity may be also essential to produce recombinants. And another explanation is that the relative positions of the EGFP sequence inserted in GFPm gene may be important [6Zhang J, Temin HM. Retrovirus recombination depends on the length of sequence identity and is not error prone. J Virol 1994; 68: 2409-14.].
Four recombination patterns might be used to produce viable recombinants during co-infection (Fig. 3 ). The exact rate of each pattern could not be determined due to the limitation of the read length (~400bp) of the 454 sequencing method used in this study, but two cross-over events were confirmed by sequence alignments (Fig. 4
). The exact rate of each pattern could not be determined due to the limitation of the read length (~400bp) of the 454 sequencing method used in this study, but two cross-over events were confirmed by sequence alignments (Fig. 4 ). The results indicated that different cross-over patterns were utilized and distinct recombination events occurred simultaneously in the viral quasispecies.
). The results indicated that different cross-over patterns were utilized and distinct recombination events occurred simultaneously in the viral quasispecies.
CONCLUSION
This study presented the detailed cross-over map of the chimeric genes in two PRRSV infectious clones by high throughput sequencing. Pattern analyses and sequence alignments indicated that different patterns may be utilized and distinct cross-over events may occur simultaneously. Our results also showed that the 20bp-conserved region is likely a cross-over hotspot, suggesting that RNA secondary structure may play a more important role for recombination in the case. This new in vitro reverse genetics system accompanied with deep sequencing creates a viable platform to study different types of recombination events and contributes to understanding of the requirements for recombination.
AUTHOR CONTRIBUTIONS
NC and RR designed the study; NC and RC performed the experiments and analyzed the data; NC, RC and RR wrote the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are base of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
This study was partially supported by China Postdoctoral Science Foundation (2016M590510) and University Science Research Project of Jiangsu Province (16KJD230001).