- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Evolving Rotaviruses, Interspecies Transmission and Zoonoses
Yashpal S. Malik1, *, Sudipta Bhat1, Parvaiz S. Dar1, Shubhankar Sircar1, Kuldeep Dhama1, Raj K. Singh1
Abstract
Evolutionary biology has become one of the imperative determinants explaining the origin of several viruses which were either identified decades back or are recognized lately using metagenomic approaches. Several notifiable emerging viruses like influenza, Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS), Ebola, Hendra, Nipah and Zika viruses have become the leading causes of epidemics and losses thereto in both human and animals. The sufferings are higher due to gastroenteritis causing viruses including Astrovirus, Calicivirus, Enterovirus, Kobuvirus Picobirnavirus, Sapelovirus, Teschovirus, and many more. Notably, the majority of the emerging viruses enclose RNA genome and these are more prone for insertions/mutation in their genome, leading to evolving viral variants. Rapidity in viral evolution becomes a big hitch in the development process of successful vaccines or antiviral. The prominent gastroenteric virus is rotavirus, which is a double-stranded RNA virus with a segmented nature of genome enabling higher reassortment events and generates unusual strains with unique genomic constellations derivative of parental rotavirus strains. Although most rotaviruses appear to be host restricted, the interspecies transmission of rotaviruses has been well documented across the globe. The nocturnal bats have been accepted harbouring many pathogenic viruses and serving as natural reservoirs. Indications are that bats can also harbour rotaviruses, and help in virus spread. The zooanthroponotic and anthropozoonotic potential of rotaviruses has significant implications for rotavirus epidemiology. Hitherto reports confirm infection of humans through rotaviruses of animal origin, exclusively via direct transmission or through gene reassortments between animal and human strain of rotaviruses. There is a need to understand the ecology and evolutionary biology of emerging rotavirus strains to design effective control programs.
Article Information
Identifiers and Pagination:
Year: 2020Volume: 14
First Page: 1
Last Page: 6
Publisher Id: TOVJ-14-1
DOI: 10.2174/1874357902014010001
Article History:
Received Date: 30/04/2019Revision Received Date: 30/11/2019
//
Acceptance Date: 05/12/2019
Electronic publication date: 18/03/2020
Collection year: 2020
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: (https://creativecommons.org/licenses/by/4.0/legalcode). This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
* Address correspondence to this author at the ICAR- Indian Veterinary Research Institute, Izatnagar, Bareilly, India; Tel: +91-581-2302777;
E-mail: malikyps@gmail.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 30-04-2019 |
Original Manuscript | Evolving Rotaviruses, Interspecies Transmission and Zoonoses | |
1. INTRODUCTION
The present world is witnessing the emergence of new microbes where the evolution rate is much higher in viruses, making them a target of substantial interest among virologists and public health specialists. Applications of modern biotechniques in deciphering the genomic data at a higher pace have added voluminous information on virus biology, although there is quiet perceptive paucity on knowing the connotation of virus evolutions. Ample data is available in support of the emergence of new viruses or virus strains which have established in uncommon host species or exhibiting species jumping, making their survival in the bionetwork conceivable. Several viral infections in humans have been tracked to have animal-origin and conversely, reverse-zoonosis is also not unusual [1Choudhary P, Minakshi P, Ranjan K, Basanti B. Zooanthroponotic transmission of rotavirus in Haryana State of Northern India. Acta Virol 2017; 61(1): 77-85.
[http://dx.doi.org/10.4149/av_2017_01_77] [PMID: 28161962] , 2Kumar N, Malik YS, Sharma K, et al. Molecular characterization of unusual bovine rotavirus A strains having high genetic relatedness with human rotavirus: Evidence for zooanthroponotic transmission. Zoonoses Public Health 2018; 65(4): 431-42.
[http://dx.doi.org/10.1111/zph.12452] [PMID: 29464925] ]. Here, the evolution in Rotavirus (RV) is addresses which has been recognized for leading gastroenteritis globally. Although, specific vaccination strategy targeting the most common group A rotavirus (RVA) has shown great success in minimizing the losses associated with the infection, the menace of infection due to other evolving RVs can not be obscured.
1.1. The Virus
Rotavirus infections are one of the leading causes of acute gastroenteritis in humans and animals throughout the world. Disproportionately they infect mostly the younger population in the developing countries, including Asian and sub-Saharan African countries. The virus belongs to the genus Rotavirus, family Reoviridae and contains 18.5kb dsRNA 11 segmented genome that makes the virus more prone to point mutations and/or genome segment reassortments. Although, this virus (RV) was shown under an electron microscope in 1973, by Ruth Bishop and co-workers (1973) in Australia, Jacob Light and Horace Hodes detected filterable agents in the faeces of children suffering with diarrhea during 1943. Subsequently, archived samples [Epizootic Diarrhea of Infant Mice (EDIM) virus in mice, Simian Agent 11 (SA11) in vervet monkey, and bovine rotavirus in diarrheic stools from calves] were confirmed to be RV [3Mebus CA, Wyatt RG, Sharpee RL, et al. Diarrhea in gnotobiotic calves caused by the reovirus-like agent of human infantile gastroenteritis. Infect Immun 1976; 14(2): 471-4.
[http://dx.doi.org/10.1128/IAI.14.2.471-474.1976] [PMID: 184047] ]. In 1974, Thomas Henry Flewett proposed the name “Rotavirus”, depicting its look as ‘Wheel’ shaped (Rota in Latin) [4Flewett TH, Woode GN. The rotaviruses. Arch Virol 1978; 57(1): 1-23.
[http://dx.doi.org/10.1007/BF01315633] [PMID: 77663] ]; and this was officially recognized after 4 years by the International Committee on Taxonomy of Viruses (ICTV) [5Matthews RE. Third report of the International Committee on Taxonomy of Viruses. Classification and nomenclature of viruses. Intervirology 1979; 12(3-5): 129-296.
[http://dx.doi.org/10.1159/000149081] [PMID: 43850] ]. At the same time, similar viruses were identified in several other animal hosts [6Woode GN, Bridger JC, Jones JM, et al. Morphological and antigenic relationships between viruses (rotaviruses) from acute gastroenteritis of children, calves, piglets, mice, and foals. Infect Immun 1976; 14(3): 804-10.
[http://dx.doi.org/10.1128/IAI.14.3.804-810.1976] [PMID: 965097] ]. Diversity in RV with the description of serotypes was first described in 1980 [7Beards GM, Pilfold JN, Thouless ME, Flewett TH. Rotavirus serotypes by serum neutralisation. J Med Virol 1980; 5(3): 231-7.
[http://dx.doi.org/10.1002/jmv.1890050307] [PMID: 6262451] ], and subsequently, the virus was successfully grown in the monkey kidneys derived cells after trypsin treatment [8Urasawa T, Urasawa S, Taniguchi K. Sequential passages of human rotavirus in MA-104 cells. Microbiol Immunol 1981; 25(10): 1025-35.
[http://dx.doi.org/10.1111/j.1348-0421.1981.tb00109.x] [PMID: 6273696] ].
1.2. Virus Classification
Based on the serological reactivity and genetic variability of group-specific antigen VP6, RVs are differentiated into 8 groups/ species/ types, designated as RVA-RVH (Rotavirus A, Rotavirus B…H, etc.) and lately, RVs identified in sheltered dogs and bats in Hungary and Serbia are proposed as new species (RVI and RVJ), although confirmation by the ICTV is pending [9Mihalov-Kovács E, Gellért Á, Marton S, et al. Candidate new rotavirus species in sheltered dogs, Hungary. Emerg Infect Dis 2015; 21(4): 660-3.
[http://dx.doi.org/10.3201/eid2104.141370] [PMID: 25811414] , 10Balato A, Scala E, Balato N, et al. Biologics that inhibit the Th17 pathway and related cytokines to treat inflammatory disorders. Expert Opin Biol Ther 2017; 17(11): 1363-74.
[http://dx.doi.org/10.1080/14712598.2017.1363884] [PMID: 28791896] ]. Since the first discovery of RVA in 1973, a discovery timeline of other RVs has been depicted in Fig. (1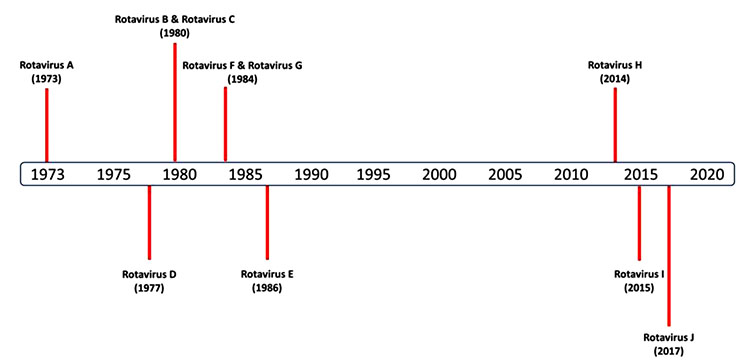 ). The first three groups, RVA, RVB and RVC, are more common pathogens of humans and various animal species. While, RVE has been reported only from pigs, and RVD, RVF and RVG are entirely found in birds [11Dhama K, Saminathan M, Karthik K, et al. Avian rotavirus enteritis - an updated review. Vet Q 2015; 35(3): 142-58.
). The first three groups, RVA, RVB and RVC, are more common pathogens of humans and various animal species. While, RVE has been reported only from pigs, and RVD, RVF and RVG are entirely found in birds [11Dhama K, Saminathan M, Karthik K, et al. Avian rotavirus enteritis - an updated review. Vet Q 2015; 35(3): 142-58.
[http://dx.doi.org/10.1080/01652176.2015.1046014] [PMID: 25917772] ].
As of now, several rotavirus group/ species have been identified in mammalian and avian host species viz. human, cattle, goats, pigs, rat, sheep, chicken and turkey, dogs, juvenile ferrets, cats, horses, non-human primates, antelope, guanaco, vicuna, bat, mouse, rabbit, giant panda, camelids to name a few [12Kusumakar AL, Savita , Malik YS, Minakshi , Prasad G. Genomic diversity among group A rotaviruses from diarrheic children, piglets, buffalo and cow calves of Madhya Pradesh. Indian J Microbiol 2010; 50(1): 83-8.
[http://dx.doi.org/10.1007/s12088-010-0016-y] [PMID: 23100812] -16Papp H, Malik YS, Farkas SL, Jakab F, Martella V, Bányai K. Rotavirus strains in neglected animal species including lambs, goats and camelids. Virusdisease 2014; 25(2): 215-22.
[http://dx.doi.org/10.1007/s13337-014-0203-2] [PMID: 25674588] ]. A classification system has recently been developed for RVA in which all the 11 genomic RNA segments are involved [17Matthijnssens J, Ciarlet M, Heiman E, et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol 2008; 82(7): 3204-19.
[http://dx.doi.org/10.1128/JVI.02257-07] [PMID: 18216098] ]. High diversity has been noted in RVA than other RVs. For example, in RVA, at least 36 G and 51 P genotypes have been recognized in both mammalian and avian species on the basis of differences in VP7 and VP4 gene sequences, respectively (https://rega.kuleuven.be/cev/ viralmetagenomics /virus-classification/rcwg). For each genome segment, various genotypes have been demarcated based on the nucleotide identity cut-off percentages. For the assessment of entire RV genomes, a nomenclature has been assigned in which the notations Gx-P[x]-Ix- Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx are used for the VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NS P3-NSP4-NSP5/6 encoding genes, respectively. This system of virus classification is an expended scheme of virus characterization and now is almost the way head to replace previous genotype-based system targeting only RV genes VP4, VP7, VP6, and NSP4. Complete genome exploration has augmented our knowledge based on the similarity between animal and human RVs, which has highlighted the significance of a common nomenclature for both animal and public health. The Rotavirus Classification Working Group (RCWG) worked to maintain, assess, and improve this system [17Matthijnssens J, Ciarlet M, Heiman E, et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol 2008; 82(7): 3204-19.
[http://dx.doi.org/10.1128/JVI.02257-07] [PMID: 18216098] ] (Table 1).
1.3. Interspecies Transmission, Reassortments and Zoonoses
The competence of virus replication in different host species is one of the important factors establishing the emergence of new viral mutants. The virus and host interact in a complex way. The virus-host restriction evolved during the coevolution of host and pathogen. The pathogen overcomes many hurdles in the replication process by adapting several molecular alterations [26Joseph U, Su YC, Vijaykrishna D, Smith GJ. The ecology and adaptive evolution of influenza A interspecies transmission. Influenza Other Respir Viruses 2017; 11(1): 74-84.
[http://dx.doi.org/10.1111/irv.12412] [PMID: 27426214] ]. Nonetheless, majorly interspecies transmission events are dead-end, but a few of the viral variants adapt to new host species and facilitate continuous spread. Emerging viruses exhibit higher inter-species transmission capacity as they possess RNA genomes and a higher rate of mutation in them facilitates adaptation to a new host.
The segmented genome of RV allows reassortment (exchange of gene segments) where unique strains emerge possessing novel genomic constellation of virus genes taken from two-parent RV strains (common in human-animal strains). Although, RVs have a preference for the selective host, on several occasions’ cross-species spread has also been foretold [17Matthijnssens J, Ciarlet M, Heiman E, et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol 2008; 82(7): 3204-19.
[http://dx.doi.org/10.1128/JVI.02257-07] [PMID: 18216098] , 27Martella V, Bányai K, Matthijnssens J, Buonavoglia C, Ciarlet M. Zoonotic aspects of rotaviruses. Vet Microbiol 2010; 140(3-4): 246-55.
[http://dx.doi.org/10.1016/j.vetmic.2009.08.028] [PMID: 19781872] -32Kattoor JJ, Saurabh S, Malik YS, et al. Unexpected detection of porcine rotavirus C strains carrying human origin VP6 gene. Vet Q 2017; 37(1): 252-61.
[http://dx.doi.org/10.1080/01652176.2017.1346849] [PMID: 28643555] ]. Lately, new genotypes have been described in a variety of animal species having multiple host origin [33Matthijnssens J, Bilcke J, Ciarlet M, et al. Rotavirus disease and vaccination: Impact on genotype diversity. Future Microbiol 2009; 4(10): 1303-16.
[http://dx.doi.org/10.2217/fmb.09.96] [PMID: 19995190] ]. The underlying mechanisms of RV diversification consist of either accumulation of single point mutations, gene recombination, rearrangement, and notably reassortment. Outwardly, RVs crossing the host species barrier remain weak to infect or spread in a new host. However, on attaining new genomic segments, these virus strains gather higher chances to infect and spread amid the new host population. A schematic representation of different animal rotavirus A (RVA) genotypes sporadically reported in humans is given in Fig. (2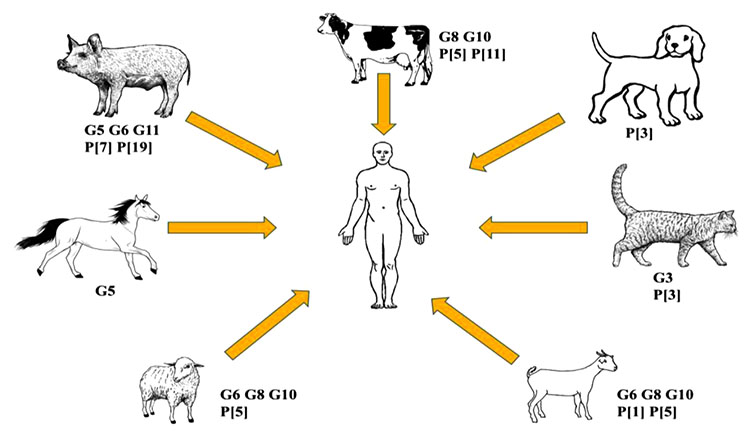 ).
).
Recently, a report from Thailand reveals porcine-like characteristics of two human RVA strains supporting the hypothesis that pigs play an important role as a source or reservoir for a novel and newly adapted porcine RVAs transmission to the human population [34Malasao R, Khamrin P, Kumthip K, Ushijima H, Maneekarn N. Complete genome sequence analysis of rare G4P[6] rotavirus strains from human and pig reveals the evidence for interspecies transmission. Infect Genet Evol 2018; 65: 357-68.
[http://dx.doi.org/10.1016/j.meegid.2018.08.019] [PMID: 30144568] ]. Alike, whole genomic G10P [14Kattoor JJ, Malik YS, Sharma K, et al. Molecular evidence of group D rotavirus in commercial broiler chicks in India. Avian Biol Res 2013; 6: 313-6.
[http://dx.doi.org/10.3184/175815513X13833072000999] ] strain from diarrhoeic child appeared to be of artiodactyl origin support bovine-to-human interspecies transmission of RVA strains [35Tacharoenmuang R, Komoto S, Guntapong R, et al. Characterization of a G10P[14] rotavirus strain from a diarrheic child in Thailand: Evidence for bovine-to-human zoonotic transmission. Infect Genet Evol 2018; 63: 43-57.
[http://dx.doi.org/10.1016/j.meegid.2018.05.009] [PMID: 29772399] ]. Notably, a phylogenetic analysis of human RV strain shown that all the 11 genes seemed of porcine origin [36Komoto S, Tacharoenmuang R, Guntapong R, et al. Identification and characterization of a human G9P[23] rotavirus strain from a child with diarrhoea in Thailand: Evidence for porcine-to-human interspecies transmission. J Gen Virol 2017; 98(4): 532-8.
[http://dx.doi.org/10.1099/jgv.0.000722] [PMID: 28382902] ], confirming adaptation of porcine RVA to humans. Complete genome analysis of porcine RVA (G5P [13Malik YS, Kumar N, Sharma K, et al. Epidemiology and genetic diversity of rotavirus strains associated with acute gastroenteritis in bovine, porcine, poultry and human population of Madhya Pradesh, Central India, 2004–2008. Adv Ani Vet Sci 2013; 2013: 111-5.]) from the Caribbean region shown that porcine-to-simian interspecies transmission of RVAs [37Navarro R, Aung MS, Cruz K, et al. Whole genome analysis provides evidence for porcine-to-simian interspecies transmission of rotavirus-A. Infect Genet Evol 2017; 49: 21-31.
[http://dx.doi.org/10.1016/j.meegid.2016.12.026] [PMID: 28039076] ]. Whole-genome analysis of human and porcine RVA (G9P [19Bridger JC. Detection by electron microscopy of caliciviruses, astroviruses and rotavirus-like particles in the faeces of piglets with diarrhoea. Vet Rec 1980; 107(23): 532-3.
[PMID: 6258286] ]) provides evidence for interspecies transmission of non-reassorted porcine RVA [38Yodmeeklin A, Khamrin P, Chuchaona W, et al. Analysis of complete genome sequences of G9P[19] rotavirus strains from human and piglet with diarrhea provides evidence for whole-genome interspecies transmission of nonreassorted porcine rotavirus. Infect Genet Evol 2017; 47: 99-108.
[http://dx.doi.org/10.1016/j.meegid.2016.11.021] [PMID: 27894992] ]. Research findings from the USA indicated the possibility of a human RVA (G14P [24Molinari BL, Lorenzetti E, Otonel RA, Alfieri AF, Alfieri AA. Species H rotavirus detected in piglets with diarrhea, Brazil, 2012. Emerg Infect Dis 2014; 20(6): 1019-22.
[http://dx.doi.org/10.3201/eid2006.130776] [PMID: 24855935] ]) strain origin by interspecies transmission and multiple reassortment events involving human, bovine and equine RVs, resulting in the introduction of some genes into the genome of simian RVs [39Mijatovic-Rustempasic S, Roy S, Teel EN, et al. Full genome characterization of the first G3P[24] rotavirus strain detected in humans provides evidence of interspecies reassortment and mutational saturation in the VP7 gene. J Gen Virol 2016; 97(2): 389-402.
[http://dx.doi.org/10.1099/jgv.0.000349] [PMID: 26590163] ].
The listing of diseases of zoonotic nature is on increase globally and several putative risk factors have been identified including urbanization, deforestation, altered population dynamics, immune pressure, mutations / genetic changes, etc. The impact of such diseases is more on socioeconomic weaker sections, particularly from developing nations of the world. The higher interaction of humans with animals including farm or per/companion animals, could also lead to interspecies transmission of such pathogens. Both anthropozoonoses and reverse-zoonoses (zoo-anthroponotic) are well noted these days in rural as well as urban areas. The RVs are not an exception and at the same time, animal RVs are considered as potential reservoirs for genetic exchange with human RV strains. Several reports confirm that RVs of animal origin infect humans via the direct spread of the animal virus strain or through gene reassortment of one or more genes of animal RV strain with a co-infecting human strain of RVs. The zoonotic potential of RVAs has important implications for understanding the human RV epidemiology. The diversity of RVs in any host species is readily refreshed by independent interspecies transmission events at any geographic location, coupled with the global transmission of the adapted virus strain. Thus, elimination of RV infections through vaccination and other human-targeted involvements is unlikely due to the large number of host species whose homologous RVs and gene pools are available in the human host [27Martella V, Bányai K, Matthijnssens J, Buonavoglia C, Ciarlet M. Zoonotic aspects of rotaviruses. Vet Microbiol 2010; 140(3-4): 246-55.
[http://dx.doi.org/10.1016/j.vetmic.2009.08.028] [PMID: 19781872] , 29Malik Y, Kumar N, Sharma K, et al. Rotavirus diarrhea in piglets: A review on epidemiology, genetic diversity and zoonotic risks. Indian J Anim Sci 2014; 84: 1035-42.]. Entirely zoonotic strains are rarely detected in surveillance studies of human RV infections [40Dóró R, Mihalov-Kovács E, Marton S, et al. Large-scale whole genome sequencing identifies country-wide spread of an emerging G9P[8] rotavirus strain in Hungary, 2012. Infect Genet Evol 2014; 28: 495-512.
[http://dx.doi.org/10.1016/j.meegid.2014.09.016] [PMID: 25239526] ]. Zoonotic transmission together with reassortment is a more efficient means for introduction of new antigen specificities to which humans are immunologically naive. During the 1990s, an introduction of G9 VP7 specificity from an animal, most likely the porcine host is an important example [41Iturriza-Gómara M, Cubitt D, Steele D, et al. Characterisation of rotavirus G9 strains isolated in the UK between 1995 and 1998. J Med Virol 2000; 61(4): 510-7.
[http://dx.doi.org/10.1002/1096-9071(200008)61:4<510::AID-JMV15>3.0.CO;2-Q] [PMID: 10897071] , 42Mijatovic-Rustempasic S, Bányai K, Esona MD, Foytich K, Bowen MD, Gentsch JR. Genome sequence based molecular epidemiology of unusual US Rotavirus A G9 strains isolated from Omaha, USA between 1997 and 2000. Infect Genet Evol 2011; 11(2): 522-7.
[http://dx.doi.org/10.1016/j.meegid.2010.11.012] [PMID: 21130184] ]. Reassortants carrying a single or few animal-origin genes within a genetic background typical of human rotavirus strains may be more transmissible in the new host. Reverse zoonosis has been well documented in rotaviruses now where human origin rotavirus strains share a few of the genetic segments with animals. Recently, human origin G1 and G9 rotavirus types are detected in bovine calves [2Kumar N, Malik YS, Sharma K, et al. Molecular characterization of unusual bovine rotavirus A strains having high genetic relatedness with human rotavirus: Evidence for zooanthroponotic transmission. Zoonoses Public Health 2018; 65(4): 431-42.
[http://dx.doi.org/10.1111/zph.12452] [PMID: 29464925] ].
1.4. Bats as Reservoirs
Bats are identified as a major link between new zoonotic pathogens identified during the last two decades, particularly. Although the majority include viral zoonoses, the pointing examples of recent past are Severe Acute Respiratory Syndrome (SAARS) and Middle East Respiratory Syndrome (MERS) coronaviruses, Nipah virus, Ebola, and Hendra virus. The close vicinity of humans and animals delivers sufficient junctures for cross-species transmission and zoonotic events to happen thereto. Bats encompass near to one-fourth of the known terrestrial mammals species (~5,500). Other than close social interfaces, bats possess several other unique allures, such as longevity, migratory activity, large and dense roosting communities, that augment their role to enable virus evolution [43Luis AD, Hayman DT, O’Shea TJ, et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: Are bats special? Proc Biol Sci 2013; 280(1756)20122753
[http://dx.doi.org/10.1098/rspb.2012.2753] [PMID: 23378666] ]. Several research finding opines towards reassortment between human, animal, and bat RVs. Furthermore, the identification of new RV species/groups in bats clearly indicates the concern of this mammalian host species in RV ecology [44Esona MD, Mijatovic-Rustempasic S, Conrardy C, et al. Reassortant group A rotavirus from straw-colored fruit bat (Eidolon helvum). Emerg Infect Dis 2010; 16(12): 1844-52.
[http://dx.doi.org/10.3201/eid1612.101089] [PMID: 21122212] ]. Notably, it is posited that several viruses utilize conserved mammalian cellular receptors and biochemical pathways available in bats, thereby transmit to other mammalian hosts, including humans [45Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: Important reservoir hosts of emerging viruses. Clin Microbiol Rev 2006; 19(3): 531-45.
[http://dx.doi.org/10.1128/CMR.00017-06] [PMID: 16847084] ]. However, still little information is available on bat biology and risk factors associated with the spillover of viruses across species. Thus, there is a need to interrogate thoroughly the potential role of wildlife animals as a passive carrier of the RVs, leading to the emergence of virus variants having a higher capacity to infect and spread in susceptible or naïve population.
1.5. Epilogue and Perspectives
Since the first description of RV in human specimens during the early 1970s, the list of host species inflicted by RVs of different types/ species has expanded many folds. The RV infections have been documented in humans, livestock farm animal species, companion animals including poultry, wildlife animals and marine species too. The virus spillage from the preferential gastrointestinal tract site to other organs of the host including the brain, liver, and spleen intimidates and raises many questions for the researchers across the globe over the past few years. The RVs are among the most unstable and frequently mutating viruses due to deficient RNA polymerase gene proofreading activity and it further accommodates the phenomenon of recombination/gene segment exchange, reproducing new genetic variants in the environment. The virus rigidity resist the adverse environment through which it passes after entering the oral route cavity and reaching the site of replication in the intestinal mucosa. Through the passage in the gut, its interaction with commensal bacteria is least explored in terms of generating co-infections and recombinations. It has been observed that the population of RNA viruses accumulates through binding to the cell wall of bacterium and upsurges the recombination of genetic material. The role of bacterial lipopolysaccharides on a number of viruses including polio has been explicated. Furthermore, reoviruses adhere to Gram’s negative and positive bacterial populations utilizing their outer layered cellular constituents and found to augment the thermal tolerance of the virus. The control of the evolving rotavirus infection in both humans and animals is dependent on regular monitoring of newly emerging viruses by effective diagnosis and typing methods [46Prasad G, Malik Y, Pandey R G. Indian J Biotechnol 2005; 4: 93-9.-48Saminathan M, Rana R, Ramakrishnan MA, Karthik K, Malik YS, Dhama K. Prevalence, diagnosis, management and control of important diseases of ruminants with special reference to Indian scenario. J Exp Biol Agric Sci 2016; 4: 338-67.
[http://dx.doi.org/10.18006/2016.4(3S).338.367] ] along with effective vaccines. Also, mixed infection along with RVs other emerging enteric dsRNA viruses have been also detected several times as opportunistic infection [49Malik YS, Chandrashekar KM, Sharma K, et al. Picobirnavirus detection in bovine and buffalo calves from foothills of Himalaya and Central India. Trop Anim Health Prod 2011; 43(8): 1475-8.
[http://dx.doi.org/10.1007/s11250-011-9834-0] [PMID: 21479844] ]. Still, there are a few primary questions to work upon, including the generation of trillions of infectious rotavirus particles per gm of stool specimen during acute infections, and mechanisms that are divulging these virus particles more contagious. A few of these queries can be resolved through exploring the viral ecological and evolutionary progressions.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
All the authors of the manuscript thank and acknowledge their respective universities and institutes.
REFERENCES
| [1] | Choudhary P, Minakshi P, Ranjan K, Basanti B. Zooanthroponotic transmission of rotavirus in Haryana State of Northern India. Acta Virol 2017; 61(1): 77-85. [http://dx.doi.org/10.4149/av_2017_01_77] [PMID: 28161962] |
| [2] | Kumar N, Malik YS, Sharma K, et al. Molecular characterization of unusual bovine rotavirus A strains having high genetic relatedness with human rotavirus: Evidence for zooanthroponotic transmission. Zoonoses Public Health 2018; 65(4): 431-42. [http://dx.doi.org/10.1111/zph.12452] [PMID: 29464925] |
| [3] | Mebus CA, Wyatt RG, Sharpee RL, et al. Diarrhea in gnotobiotic calves caused by the reovirus-like agent of human infantile gastroenteritis. Infect Immun 1976; 14(2): 471-4. [http://dx.doi.org/10.1128/IAI.14.2.471-474.1976] [PMID: 184047] |
| [4] | Flewett TH, Woode GN. The rotaviruses. Arch Virol 1978; 57(1): 1-23. [http://dx.doi.org/10.1007/BF01315633] [PMID: 77663] |
| [5] | Matthews RE. Third report of the International Committee on Taxonomy of Viruses. Classification and nomenclature of viruses. Intervirology 1979; 12(3-5): 129-296. [http://dx.doi.org/10.1159/000149081] [PMID: 43850] |
| [6] | Woode GN, Bridger JC, Jones JM, et al. Morphological and antigenic relationships between viruses (rotaviruses) from acute gastroenteritis of children, calves, piglets, mice, and foals. Infect Immun 1976; 14(3): 804-10. [http://dx.doi.org/10.1128/IAI.14.3.804-810.1976] [PMID: 965097] |
| [7] | Beards GM, Pilfold JN, Thouless ME, Flewett TH. Rotavirus serotypes by serum neutralisation. J Med Virol 1980; 5(3): 231-7. [http://dx.doi.org/10.1002/jmv.1890050307] [PMID: 6262451] |
| [8] | Urasawa T, Urasawa S, Taniguchi K. Sequential passages of human rotavirus in MA-104 cells. Microbiol Immunol 1981; 25(10): 1025-35. [http://dx.doi.org/10.1111/j.1348-0421.1981.tb00109.x] [PMID: 6273696] |
| [9] | Mihalov-Kovács E, Gellért Á, Marton S, et al. Candidate new rotavirus species in sheltered dogs, Hungary. Emerg Infect Dis 2015; 21(4): 660-3. [http://dx.doi.org/10.3201/eid2104.141370] [PMID: 25811414] |
| [10] | Balato A, Scala E, Balato N, et al. Biologics that inhibit the Th17 pathway and related cytokines to treat inflammatory disorders. Expert Opin Biol Ther 2017; 17(11): 1363-74. [http://dx.doi.org/10.1080/14712598.2017.1363884] [PMID: 28791896] |
| [11] | Dhama K, Saminathan M, Karthik K, et al. Avian rotavirus enteritis - an updated review. Vet Q 2015; 35(3): 142-58. [http://dx.doi.org/10.1080/01652176.2015.1046014] [PMID: 25917772] |
| [12] | Kusumakar AL, Savita , Malik YS, Minakshi , Prasad G. Genomic diversity among group A rotaviruses from diarrheic children, piglets, buffalo and cow calves of Madhya Pradesh. Indian J Microbiol 2010; 50(1): 83-8. [http://dx.doi.org/10.1007/s12088-010-0016-y] [PMID: 23100812] |
| [13] | Malik YS, Kumar N, Sharma K, et al. Epidemiology and genetic diversity of rotavirus strains associated with acute gastroenteritis in bovine, porcine, poultry and human population of Madhya Pradesh, Central India, 2004–2008. Adv Ani Vet Sci 2013; 2013: 111-5. |
| [14] | Kattoor JJ, Malik YS, Sharma K, et al. Molecular evidence of group D rotavirus in commercial broiler chicks in India. Avian Biol Res 2013; 6: 313-6. [http://dx.doi.org/10.3184/175815513X13833072000999] |
| [15] | Mondal A, Sharma K, Malik YS, Joardar SN. Detection of group a rotavirus in faeces of diarrhoeic bovine porcine and human population from eastern India by reverse transcriptase–polymerase chain reaction. Adv Ani Vet Sci 2013; 1: 18-9. |
| [16] | Papp H, Malik YS, Farkas SL, Jakab F, Martella V, Bányai K. Rotavirus strains in neglected animal species including lambs, goats and camelids. Virusdisease 2014; 25(2): 215-22. [http://dx.doi.org/10.1007/s13337-014-0203-2] [PMID: 25674588] |
| [17] | Matthijnssens J, Ciarlet M, Heiman E, et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol 2008; 82(7): 3204-19. [http://dx.doi.org/10.1128/JVI.02257-07] [PMID: 18216098] |
| [18] | Bishop RF, Davidson GP, Holmes IH, Ruck BJ. Virus particles in epithelial cells of duodenal mucosa from children with acute non-bacterial gastroenteritis. Lancet 1973; 2(7841): 1281-3. [http://dx.doi.org/10.1016/S0140-6736(73)92867-5] [PMID: 4127639] |
| [19] | Bridger JC. Detection by electron microscopy of caliciviruses, astroviruses and rotavirus-like particles in the faeces of piglets with diarrhoea. Vet Rec 1980; 107(23): 532-3. [PMID: 6258286] |
| [20] | Saif LJ, Bohl EH, Theil KW, Cross RF, House JA. Rotavirus-like, calicivirus-like, and 23-nm virus-like particles associated with diarrhea in young pigs. J Clin Microbiol 1980; 12(1): 105-11. [http://dx.doi.org/10.1128/JCM.12.1.105-111.1980] [PMID: 6252238] |
| [21] | Bergeland M, McAdaragh J, Stotz I. Rotaviral enteritis in turkey poults. Proceedings of Western Poultry Disease Conference and Poultry Health Symposium: Cooperative Extension 1977; 129-30. |
| [22] | Chasey D, Bridger JC, McCrae MA. A new type of atypical rotavirus in pigs. Arch Virol 1986; 89(1-4): 235-43. [http://dx.doi.org/10.1007/BF01309892] [PMID: 3013130] |
| [23] | McNulty MS, Todd D, Allan GM, McFerran JB, Greene JA. Epidemiology of rotavirus infection in broiler chickens: Recognition of four serogroups. Arch Virol 1984; 81(1-2): 113-21. [http://dx.doi.org/10.1007/BF01309301] [PMID: 6331344] |
| [24] | Molinari BL, Lorenzetti E, Otonel RA, Alfieri AF, Alfieri AA. Species H rotavirus detected in piglets with diarrhea, Brazil, 2012. Emerg Infect Dis 2014; 20(6): 1019-22. [http://dx.doi.org/10.3201/eid2006.130776] [PMID: 24855935] |
| [25] | Bányai K, Kemenesi G, Budinski I, et al. Candidate new rotavirus species in Schreiber’s bats, Serbia. Infect Genet Evol 2017; 48: 19-26. [http://dx.doi.org/10.1016/j.meegid.2016.12.002] [PMID: 27932285] |
| [26] | Joseph U, Su YC, Vijaykrishna D, Smith GJ. The ecology and adaptive evolution of influenza A interspecies transmission. Influenza Other Respir Viruses 2017; 11(1): 74-84. [http://dx.doi.org/10.1111/irv.12412] [PMID: 27426214] |
| [27] | Martella V, Bányai K, Matthijnssens J, Buonavoglia C, Ciarlet M. Zoonotic aspects of rotaviruses. Vet Microbiol 2010; 140(3-4): 246-55. [http://dx.doi.org/10.1016/j.vetmic.2009.08.028] [PMID: 19781872] |
| [28] | Malik YS, Sharma K, Vaid N, et al. Frequency of group A rotavirus with mixed G and P genotypes in bovines: Predominance of G3 genotype and its emergence in combination with G8/G10 types. J Vet Sci 2012; 13(3): 271-8. [http://dx.doi.org/10.4142/jvs.2012.13.3.271] [PMID: 23006956] |
| [29] | Malik Y, Kumar N, Sharma K, et al. Rotavirus diarrhea in piglets: A review on epidemiology, genetic diversity and zoonotic risks. Indian J Anim Sci 2014; 84: 1035-42. |
| [30] | Malik YS, Kumar N, Sharma K, et al. Multispecies reassortant bovine rotavirus strain carries a novel simian G3-like VP7 genotype. Infect Genet Evol 2016; 41: 63-72. [http://dx.doi.org/10.1016/j.meegid.2016.03.023] [PMID: 27033751] |
| [31] | Ghosh S, Navarro R, Malik YS, Willingham AL, Kobayashi N. Whole genomic analysis of a porcine G6P[13] rotavirus strain. Vet Microbiol 2015; 180(3-4): 286-98. [http://dx.doi.org/10.1016/j.vetmic.2015.09.017] [PMID: 26454565] |
| [32] | Kattoor JJ, Saurabh S, Malik YS, et al. Unexpected detection of porcine rotavirus C strains carrying human origin VP6 gene. Vet Q 2017; 37(1): 252-61. [http://dx.doi.org/10.1080/01652176.2017.1346849] [PMID: 28643555] |
| [33] | Matthijnssens J, Bilcke J, Ciarlet M, et al. Rotavirus disease and vaccination: Impact on genotype diversity. Future Microbiol 2009; 4(10): 1303-16. [http://dx.doi.org/10.2217/fmb.09.96] [PMID: 19995190] |
| [34] | Malasao R, Khamrin P, Kumthip K, Ushijima H, Maneekarn N. Complete genome sequence analysis of rare G4P[6] rotavirus strains from human and pig reveals the evidence for interspecies transmission. Infect Genet Evol 2018; 65: 357-68. [http://dx.doi.org/10.1016/j.meegid.2018.08.019] [PMID: 30144568] |
| [35] | Tacharoenmuang R, Komoto S, Guntapong R, et al. Characterization of a G10P[14] rotavirus strain from a diarrheic child in Thailand: Evidence for bovine-to-human zoonotic transmission. Infect Genet Evol 2018; 63: 43-57. [http://dx.doi.org/10.1016/j.meegid.2018.05.009] [PMID: 29772399] |
| [36] | Komoto S, Tacharoenmuang R, Guntapong R, et al. Identification and characterization of a human G9P[23] rotavirus strain from a child with diarrhoea in Thailand: Evidence for porcine-to-human interspecies transmission. J Gen Virol 2017; 98(4): 532-8. [http://dx.doi.org/10.1099/jgv.0.000722] [PMID: 28382902] |
| [37] | Navarro R, Aung MS, Cruz K, et al. Whole genome analysis provides evidence for porcine-to-simian interspecies transmission of rotavirus-A. Infect Genet Evol 2017; 49: 21-31. [http://dx.doi.org/10.1016/j.meegid.2016.12.026] [PMID: 28039076] |
| [38] | Yodmeeklin A, Khamrin P, Chuchaona W, et al. Analysis of complete genome sequences of G9P[19] rotavirus strains from human and piglet with diarrhea provides evidence for whole-genome interspecies transmission of nonreassorted porcine rotavirus. Infect Genet Evol 2017; 47: 99-108. [http://dx.doi.org/10.1016/j.meegid.2016.11.021] [PMID: 27894992] |
| [39] | Mijatovic-Rustempasic S, Roy S, Teel EN, et al. Full genome characterization of the first G3P[24] rotavirus strain detected in humans provides evidence of interspecies reassortment and mutational saturation in the VP7 gene. J Gen Virol 2016; 97(2): 389-402. [http://dx.doi.org/10.1099/jgv.0.000349] [PMID: 26590163] |
| [40] | Dóró R, Mihalov-Kovács E, Marton S, et al. Large-scale whole genome sequencing identifies country-wide spread of an emerging G9P[8] rotavirus strain in Hungary, 2012. Infect Genet Evol 2014; 28: 495-512. [http://dx.doi.org/10.1016/j.meegid.2014.09.016] [PMID: 25239526] |
| [41] | Iturriza-Gómara M, Cubitt D, Steele D, et al. Characterisation of rotavirus G9 strains isolated in the UK between 1995 and 1998. J Med Virol 2000; 61(4): 510-7. [http://dx.doi.org/10.1002/1096-9071(200008)61:4<510::AID-JMV15>3.0.CO;2-Q] [PMID: 10897071] |
| [42] | Mijatovic-Rustempasic S, Bányai K, Esona MD, Foytich K, Bowen MD, Gentsch JR. Genome sequence based molecular epidemiology of unusual US Rotavirus A G9 strains isolated from Omaha, USA between 1997 and 2000. Infect Genet Evol 2011; 11(2): 522-7. [http://dx.doi.org/10.1016/j.meegid.2010.11.012] [PMID: 21130184] |
| [43] | Luis AD, Hayman DT, O’Shea TJ, et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: Are bats special? Proc Biol Sci 2013; 280(1756)20122753 [http://dx.doi.org/10.1098/rspb.2012.2753] [PMID: 23378666] |
| [44] | Esona MD, Mijatovic-Rustempasic S, Conrardy C, et al. Reassortant group A rotavirus from straw-colored fruit bat (Eidolon helvum). Emerg Infect Dis 2010; 16(12): 1844-52. [http://dx.doi.org/10.3201/eid1612.101089] [PMID: 21122212] |
| [45] | Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: Important reservoir hosts of emerging viruses. Clin Microbiol Rev 2006; 19(3): 531-45. [http://dx.doi.org/10.1128/CMR.00017-06] [PMID: 16847084] |
| [46] | Prasad G, Malik Y, Pandey R G. Indian J Biotechnol 2005; 4: 93-9. |
| [47] | Basera SS, Singh R, Vaid N, Sharma K, Chakravarti S, Malik YP. Detection of rotavirus infection in bovine calves by RNA-PAGE and RT-PCR. Indian J Virol 2010; 21(2): 144-7. [http://dx.doi.org/10.1007/s13337-010-0017-9] [PMID: 23637494] |
| [48] | Saminathan M, Rana R, Ramakrishnan MA, Karthik K, Malik YS, Dhama K. Prevalence, diagnosis, management and control of important diseases of ruminants with special reference to Indian scenario. J Exp Biol Agric Sci 2016; 4: 338-67. [http://dx.doi.org/10.18006/2016.4(3S).338.367] |
| [49] | Malik YS, Chandrashekar KM, Sharma K, et al. Picobirnavirus detection in bovine and buffalo calves from foothills of Himalaya and Central India. Trop Anim Health Prod 2011; 43(8): 1475-8. [http://dx.doi.org/10.1007/s11250-011-9834-0] [PMID: 21479844] |