- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
In Vivo Fluorescent Labeling of Tumor Cells with the HaloTag® Technology
Jen-Chieh Tseng*, 1, Hélène A Benink3, Mark G McDougall4, Isabel Chico-Calero2, Andrew L Kung1 , 2
Abstract
Many fluorescent sensors are currently available for in vitro bio-physiological microscopic imaging. The ability to label cells in living animals with these fluorescent sensors would help translate some of these assays into in vivo applications. To achieve this goal, the first step is to establish a method for selectively labeling target cells with exogenous fluorophores. Here we tested whether the HaloTag® protein tagging system provides specific labeling of xenograft tumors in living animals. After systemic delivery of fluorophore-conjugated ligands, we performed whole animal planar fluorescent imaging to determine uptake in tag-expressing HCT116 xenografts. Our results demonstrate that HaloTag ligands containing red or near-infrared fluorophores have enhanced tumor uptake and are suitable for non-invasive in vivo imaging. Our proof-of-concept results establish feasibility for using HaloTag technology for bio-physiological imaging in living animals.
Article Information
Identifiers and Pagination:
Year: 2012Volume: 6
Issue: Suppl 1
First Page: 48
Last Page: 54
Publisher Id: CCGTM-6-48
DOI: 10.2174/1875397301206010048
Article History:
Received Date: 30/3/2012Revision Received Date: 17/5/2012
Acceptance Date: 18/5/2012
Electronic publication date: 20/9/2012
Collection year: 2012
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Lurie Family Imaging Center, Dana-Farber Cancer Institute 450 Brookline Avenue, DD 109, Boston, MA 02215, USA; Tel: 617-582-8970; Fax: 617-582-8973; E-mail: jen-chieh_tseng@dfci.harvard.edu
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 30-3-2012 |
Original Manuscript | In Vivo Fluorescent Labeling of Tumor Cells with the HaloTag® Technology | |
INTRODUCTION
The major goal of molecular imaging is to non-invasively monitor bio-physiological activities in living organisms. Recent advancements in fluorescent probe development have made it possible to monitor bio-physiological changes, such as pH [1Whitaker JE, Haugland RP, Prendergast FG. Spectral and photophysical studies of benzo[c]xanthene dyes: dual emission pH sensors Anal Biochem 1991; 194: 330-44.], ions [2Wulff H, Castle NA, Pardo LA. Voltage-gated potassium channels as therapeutic targets Nat Rev Drug Discov 2009; 8: 982-1001.], redox state [3Price M, Reiners JJ, Santiago AM, Kessel D. Monitoring singlet oxygen and hydroxyl radical formation with fluorescent probes during photodynamic therapy Photochem Photobiol 2009; 85: 1177-81.], and membrane potential [4Bunting JR, Phan TV, Kamali E, Dowben RM. Fluorescent cationic probes of mitochondria.Metrics and mechanism of interaction Biophys J 1989; 56: 979-3.] within living cells using microscopy. Extending the use of such bio-physiological sensors into animal models would be of great interest for biomedical research and drug development.
A common method used to enable fluorescence imaging of cells is via ectopic expression of recombinant fluorescent proteins, such as green-fluorescent protein (GFP), red-fluorescent protein (RFP) and their variants [5Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function Science 2006; 312: 217-4.]. Specific expression of fluorescent proteins can be achieved via tissue-specific promoters or gene delivery vector systems, and their expression can be readily monitored both by microscopy and with non-invasive imaging in live animals. However, it is difficult to develop physiological sensors based on fluorescent proteins [6Palmer AE, Qin Y, Park JG, McCombs JE. Design and application of genetically encoded biosensors Trends Biotechnol 2011; 29: 144-52.]. The deeply buried fluorescent core within the protein makes it challenging to couple fluorescence with sensors of external changes without adversely affecting the fluorescence efficiency of the protein. Another common strategy for in vivo fluorescence imaging is to use fluorophore-conjugated antibodies for cell labeling [7Ballou B, Fisher GW, Hakala TR, Farkas DL. Tumor detection and visualization using cyanine fluorochrome-labeled antibodies Biotechnol Prog 1997; 13: 649-58.]. However, relatively weak binding affinities and rapid probe clearance make antibody-fluorophore conjugates unsuitable for stable and long-term staining. In addition, antibody-based probes are intrinsically larger than small, membrane-permeable fluorophores, and thus cannot be used to monitor intracellular processes.
The HaloTag technology provides a promising alternative for in vivo fluorescent labeling. Developed based on a modified bacterial haloalkane dehalogenase, the protein tag is capable of covalent linkage to its synthetic ligands [8Los GV, Wood K. The HaloTag: a novel technology for cell imaging and protein analysis Methods Mol Biol 2007; 356: 195-208.]. This labeling is irreversible and based upon specific interaction with the tag protein. More importantly, because the ligand is comprised of a small chloroalkane linker (responsible for interaction with the tag) and a functional moiety, novel ligands are easily made by attaching this linker to a variety of physiological sensors [9Algar WR, Prasuhn DE, Stewart MH, et al. The controlled display of biomolecules on nanoparticles: a challenge suited to bioorthogonal chemistry Bioconjug Chem 2011; 22: 825-58.]. Further, compared with conventional fluorescent proteins and antibody-conjugates, these synthetic fluorescent ligands are smaller and can be membrane permeable. Of particular interest to the present study, monitoring of intracellular pH changes has been previous reported using a HaloTag ligand that contains a pH sensing module SNARF-1™ [10Benink H, McDougall M, Klaubert D, Los G. Direct pH measurements by using subcellular targeting of 5(and 6-) carboxyseminaphthorhodafluor in mammalian cells Biotechniques 2009; 47: 769-4.]. To further investigate the possibility of translating the HaloTag technology from in vitro cell-based to in vivo animal applications, we performed fluorescent labeling of tag-expressing cancer cells in living animals which are known to be exposed to an acidic pH eviroment during tumorogenesis. Using a HaloTag-expressing HCT116 human colon cancer xenograft model and either a NIR standard fluorescent ligand or the pH sensing SNARF-1™ ligand, we provide proof-of-principle for tag-specific labeling after systemic delivery of fluorescent ligands.
MATERIALS AND METHODS
Fluorescence Spectrum Unmixing of SNARF HaloTag Ligand at Different pH
The SNARF HaloTag ligand is the parent structure of the previously described AcSNARF(06)Cl [10Benink H, McDougall M, Klaubert D, Los G. Direct pH measurements by using subcellular targeting of 5(and 6-) carboxyseminaphthorhodafluor in mammalian cells Biotechniques 2009; 47: 769-4.]. In brief, SNA-RF-1™ (in the form of carboxylic acid, acetate, succinimidyl ester, Invitrogen, Carlsbad, CA) was reacted with the HaloTag chloroalkane amine linker, NH2(CH2CH2O)6(CH2)6Cl, to form SNARF-(O6)-Cl, which is referred to as SNARF ligand throughout the manuscript. To determine the fluorescence properties of this ligand at various pH, we performed fluorescence imaging in a black 96-well plate using 50 µg of the ligand in 300 µL of PBS, with pH ranging from pH 5.4 to pH 10.8.
Using the IVIS Spectrum system (Caliper Life Sciences, Hopkinton MA), the plate was sequentially imaged with a fixed excitation filter at 535 nm to determine the optimal emission in the range from 580 nm to 840 nm (1 sec, F-stop = 2, small binning). The acidic and basic SNARF fluorescence spectra were reconstructed using spectral unmixing (Living Image Software Version 4.2, Caliper Life Sciences, Hopkinton MA).
Cellular Labeling Using SNARF HaloTag Ligand in Vitro
A pCINeo-HT7 plasmid containing a cytosolic HaloTag protein expression cassette was stably transfected into HCT116 human colon cancer cells. The HaloTag-expressing cells (HCT116-HT) were cultured in DMEM with 10% FBS under selection with G418 (1 mg/mL). For in vitro cell labeling, HCT116-HT and parental HCT116 cells were incubated in culture media containing 20 µM of SNARF-1 ligand at 37°C for 15 min. After three washes with PBS, cells were incubated with fresh media for 30 min prior to imaging on a Nikon Eclipse Ti inverted fluorescence microscope.
In Vivo Spectral Characterization of SNARF HaloTag Ligand
In order to reconstruct the acidic and basic SNARF fluorescence spectra in vivo, NCr nude mice were subcutaneously inoculated with 3 × 106 HCT116 (left flank) or HCT116–HT (right flank) cells. Animals were fed an alfalfa-free diet to reduce background autofluorescence. After two weeks, each tumor-bearing mouse received a single intravenous injection of SNARF ligand (1 mg dissolved in 100 µl of 20% DMSO and 80% PBS). The day after injection, sequential fluorescent images were acquired using the IVIS Spectrum system. With a fixed Ex 535 nm filter, we performed emission scanning ranging from 580 to 800 nm (1 sec, small binning and F-stop = 2). Using the Living Image software package, the tissue autofluorescence and the SNARF fluorescence were unmixed and spectra for both components were reconstructed. We determined the optimal excitation wavelength by scanning from 535 to 640 nm, with a fixed 700 nm emission filter (1 sec, F-stop = 2 and small binning).
In Vivo Uptake of Fluorescent Ligands in HaloTag-expressing Tumor Xenografts
NCr nude mice with tumor xenografts (left: HCT116; right: HCT116-HT) received intravenous injections of 1 mg SNARF ligand (in 100 µl of 20% DMSO and 80% PBS). The ligand distribution was subsequently determined by fluorescence imaging 1 h (day 0), 1 day and 2 days after probe injection with an Ex 640/ Em 700 filter pair (1 sec, F-stop = 2 and small binning). Statistical analysis was performed using Prism (Version 5.02, GraphPad, La Jolla, CA).
To generate the IRDye800 HaloTag ligand, HaloTag chloroalkane amine linker, NH2(CH2CH2O)4(CH2)6Cl, was reacted with the IRDye800CW™ NHS ester (LI-COR, Lincoln NE) to form IRDye800-(O4)-Cl, which is referred to as IRDye800 ligand throughout the study. For in vivo tumor labeling with the IRDye800 ligand, NCr nude mice with tumor xenografts on the flanks (left: HCT116; right: HCT116-HT) received intravenous injections of 0.25 mg IRDye800 ligand (in 200 µl of 10% DMSO and 90% PBS). Ligand distribution was determined 2 h (day 0), 1 day, 2 days and 3 days after probe injection, using an Ex 745 nm/Em 800 filter set (1 sec, F-stop = 2 and small binning). Statistical analysis was performed using Prism.
All animal studies were performed under protocols approved by the DFCI ACUC.
RESULTS
The SNARF-1™ conjugated ligand was first validated in vitro as a pH sensor. SNARF ligand was dissolved in PBS at various pH (from acidic pH 5.4 to basic pH 10.8) in a black opaque 96-well plate (Fig. 1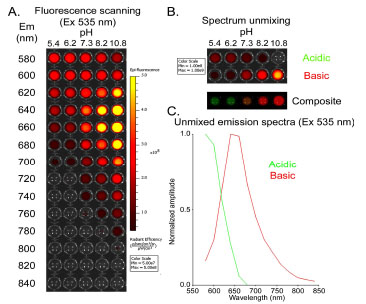 ). With a fixed excitation wavelength at 535 nm, we scanned the plate for optimal emission settings (from 580 nm to 840 nm) in an attempt to distinguish the acidic and the basic forms of SNARF ligand (Fig. 1A
). With a fixed excitation wavelength at 535 nm, we scanned the plate for optimal emission settings (from 580 nm to 840 nm) in an attempt to distinguish the acidic and the basic forms of SNARF ligand (Fig. 1A ). Using spectral unmixing, we reconstructed two fluorescence emission components corresponding to the acidic and basic spectra of the SNARF ligand (Fig. 1B
). Using spectral unmixing, we reconstructed two fluorescence emission components corresponding to the acidic and basic spectra of the SNARF ligand (Fig. 1B ). The reconstructed spectra indicate that the maximal emission is ~580 nm for acidic SNARF, and ~640 nm for basic dye (Fig. 1C
). The reconstructed spectra indicate that the maximal emission is ~580 nm for acidic SNARF, and ~640 nm for basic dye (Fig. 1C ).
).
To assess HaloTag function in vivo, we generated HaloTag-expressing xenograft tumors. Human HCT116 colon cancer cells were stably transfected with a cytosolic HaloTag expression plasmid to form the stable cell line HCT116-HT. After labeling with SNARF ligand in vitro, fluorescence was observed in the HCT116-HT cells, but not parental HCT116 cells (Fig. 2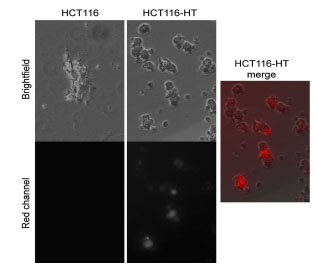 ).
).
We next tested if the SNARF ligand can label HCT116-HT tumors in vivo after systemic administration. Athymic nude mice were simultaneously inoculated with HCT116 (left) and HCT116–HT (right) cells. After establishment of subcutaneous tumors, mice were intravenously injected with SNARF ligand. Using a 535 nm excitation filter, we performed sequential emission scanning 24 h after probe injection covering the emission range from 580 to 800 nm, (Fig. 3A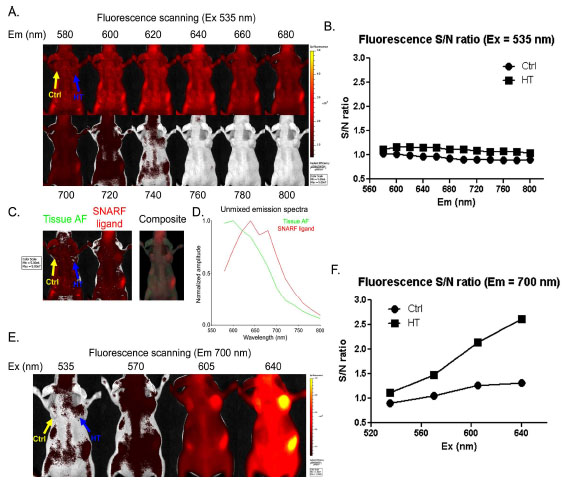 ). Unfortunately, there was significant interference from tissue autofluorescence at this excitation wavelength, resulting in low S/N ratios in tumors (Fig. 3B
). Unfortunately, there was significant interference from tissue autofluorescence at this excitation wavelength, resulting in low S/N ratios in tumors (Fig. 3B ). Although spectral unmixing of the sequential imaging data was capable of distinguishing SNARF fluorescence from tissue autofluorescence (Fig. 3C
). Although spectral unmixing of the sequential imaging data was capable of distinguishing SNARF fluorescence from tissue autofluorescence (Fig. 3C ), the reconstructed SNARF spectrum revealed considerable overlap with autofluorescence (Fig. 3D
), the reconstructed SNARF spectrum revealed considerable overlap with autofluorescence (Fig. 3D ). Thus, the optimal settings for in vitro imaging (Ex 535 /Em 580 for acidic and Ex 535 /Em 640) were confounded by significant autofluorescence in vivo.
). Thus, the optimal settings for in vitro imaging (Ex 535 /Em 580 for acidic and Ex 535 /Em 640) were confounded by significant autofluorescence in vivo.
To decrease autofluorescence and enhance SNARF signal detection in vivo, we investigated higher excitation wavelengths coupled with a 700 nm emission filter. Scanning excitation wavelengths spanning 535-640 nm revealed that a 640 nm excitation resulted in intense SNARF fluorescence and better S/N ratios in tumors (Fig. 3E , F
, F ). In addition to decreased autoflourescence, the more red-shifted excitation and emission wavelengths enable better tissue penetration.
). In addition to decreased autoflourescence, the more red-shifted excitation and emission wavelengths enable better tissue penetration.
Using this optimized in vivo setting (Ex 640 nm/ Em 700 nm), we were able to determine the kinetics of SNARF ligand uptake and retention in tumors (Fig. 4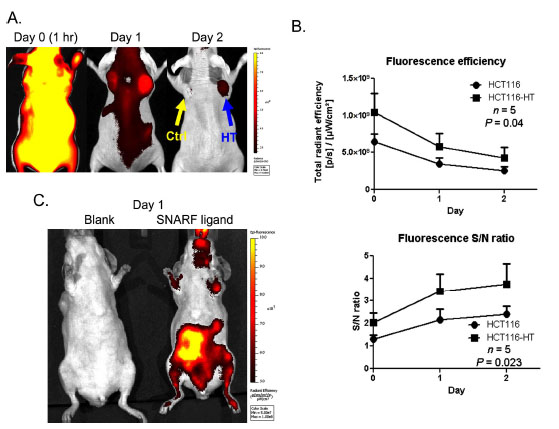 ). After systemic probe injection, higher SNARF uptake was observed in the HaloTag-expressing tumor throughout the course of imaging (Fig. 4A
). After systemic probe injection, higher SNARF uptake was observed in the HaloTag-expressing tumor throughout the course of imaging (Fig. 4A ) and HaloTag-associated signals lasted for at least 2 days after injection (Fig. 4B
) and HaloTag-associated signals lasted for at least 2 days after injection (Fig. 4B ). Using the fluorescent signal in the neck region as reference, HCT116-HT tumors exhibit above background fluorescence (Fig. 4B
). Using the fluorescent signal in the neck region as reference, HCT116-HT tumors exhibit above background fluorescence (Fig. 4B ). Although it was determined that tag-expressing tumors had statistically significant signal-to-noise over background (p<0.05), we did observe background SNARF signal in parental HCT116 tumors. In addition to uptake in tumors, above-background signal was observed in the gastrointestinal tract 1 day after probe injection (Fig. 4C
). Although it was determined that tag-expressing tumors had statistically significant signal-to-noise over background (p<0.05), we did observe background SNARF signal in parental HCT116 tumors. In addition to uptake in tumors, above-background signal was observed in the gastrointestinal tract 1 day after probe injection (Fig. 4C ).
).
To further enhance sensitivity, we further explored the possibility of using a near-infrared ligand to improve tissue penetration. A HaloTag ligand was conjugated with IRdye 800CW™ (IRdye800 ligand) which has near-infrared spectral properties (ex 795 nm, em 820 nm) ideally suited for in vivo imaging. After systemic injection of ligand, we observed intense fluorescence throughout the body and tumors 2 h after injection (Fig 5A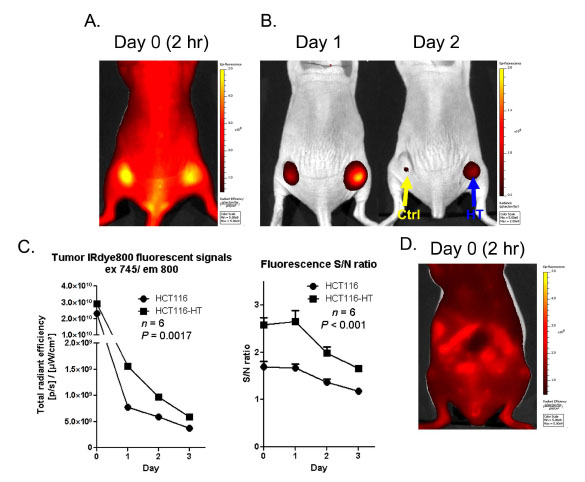 ). At this early stage, both parental HCT116 and HaloTag-expressing HCT116-HT tumors had similar fluorescence levels as a result of diffuse vascular biodistribution. In a fashion similar to the imaging results obtained from the SNARF ligand, with time we observed selective uptake and retention of the IRdye800 ligand in tag-expressing tumors for at least 3 days after probe injection (Fig. 5B
). At this early stage, both parental HCT116 and HaloTag-expressing HCT116-HT tumors had similar fluorescence levels as a result of diffuse vascular biodistribution. In a fashion similar to the imaging results obtained from the SNARF ligand, with time we observed selective uptake and retention of the IRdye800 ligand in tag-expressing tumors for at least 3 days after probe injection (Fig. 5B and C
and C ). In addition, HCT116-HT tumors exhibited signal-to-noise ratios that were significantly higher than HCT116 parental control tumors (p<0.001) (Fig. 5C
). In addition, HCT116-HT tumors exhibited signal-to-noise ratios that were significantly higher than HCT116 parental control tumors (p<0.001) (Fig. 5C ). Similar to the SNARF ligand, we observed above-background signal associated with the gastrointestinal tract as early as 2 h after IRdye800 ligand injection (Fig. 5D
). Similar to the SNARF ligand, we observed above-background signal associated with the gastrointestinal tract as early as 2 h after IRdye800 ligand injection (Fig. 5D ).
).
DISCUSSION
In this study, we provide proof-of-principle for using HaloTag-mediated fluorescence labeling of tumor cells in living animals. We demonstrate that flourophore-conjugated ligands are well tolerated after systemic delivery and there is selective fluorescence uptake in HaloTag-expressing tumors in vivo. Although the uptake was enhanced in HaloTag-expressing tumors, we did observe some non-selective signal in tag-negative tumors, likely due to EPR effects (enhanced permeability and retention), which is commonly found in rapidly growing tumors. Many fluorophores demonstrate considerable EPR effects after systemic delivery, resulting in non-specific trapping within tumor stroma [11Cheng Z, Levi J, Xiong Z, et al. Near-infrared fluorescent deoxyglucose analogue for tumor optical imaging in cell culture and living mice Bioconjug Chem 2006; 17: 662-9., 12Tseng JC, Wang Y, Banerjee P, Kung AL. Incongruity of imaging using fluorescent 2-DG conjugates compared to (18)F-FDG in preclinical cancer models Mol Imaging Biol 2012. [Epub ahead of print]]. Further, we also observed some non-specific fluorescent signal in the abdominal region. We believe that this is likely due to the ligands’ hydrophobicity/hydrophilicity, as well as other parameters (e.g. pKa, electric charge, aromatic size, and amphiphilic index) which are known to influence pharmacokinetics and clearance in animal models. Further, it is also possible that the conjugated ligand may interact with tissue components in a non-selective manner in the animal. As all ligands for HaloTag contain a chloroalkane in additional to the functional moiety, we plan to modify this region in future studies to establish a structure-activity relationship for ligand behavior in vivo.
Labeling of cells or tissues with exogenously delivered fluorophores is the first step toward successful in vivo physiological fluorescence imaging. Several hurdles need to be overcome in order to make this technology more practical for in vivo use. In general, compared with bioluminescence methods, fluorescence imaging methods have higher autofluorescence and thus lower signal-to-noise ratios. Spectral shifts resulting from microenvironmental influences (e.g., pH, protein binding, and pigments) also impact in vivo fluorescence imaging. In addition to the ligand linker modifications mention above, another way to circumvent such issues is to develop fluorescent probes that are fluorescently inert but can be effectively activated by specific intracellular biological activities. Such “gain-of-function” fluorescence sensors may significantly reduce non-specific noise, allowing for improved sensitivity and specificity for in vivo imaging.
In the current study, we tested the uptake of fluorescent ligands by a HaloTag protein which is small enough to reside throughout the interior of cell. However, fusion of HaloTag to proteins with precise subcellular localizations could be used to monitor biophysiological changes in particular sub-cellular structures of interest, such as the nucleus, mitochondria or plasma membrane. Fusion of the tag with proteins of better stability and longer half-life could prolong fluorescent signal retention with target cells. Another potential application is to use HaloTag fusion proteins, in conjunction with bioluminescence, for in vivo BRET imaging [13Xia Z, Rao J. Biosensing and imaging based on bioluminescence resonance energy transfer Curr Opin Biotechnol 2009; 20: 37-44.]. With the current success and wide adaptation of the HaloTag technology for in vitro applications, our results suggest that the technology also has potential to become a versatile platform for in vivo imaging.
CONFLICT OF INTEREST
Some authors on this study are employees of Promega Corporation or Promega Biosciences, LLC.
ACKNOWLEDGEMENT
This work was supported by the Nancy Lurie Marks Foundation.







