- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Minor Variant Detection at Different Template Concentrations in HIV-1 Phenotypic and Genotypic Tropism Testing
Ina Vandenbroucke*, §, Veerle Van Eygen§, Evelien Rondelez, Hans Vermeiren, Kurt Van Baelen, Lieven J Stuyver
Abstract
The clinical trials of maraviroc showed that treatment failure was mostly associated with lack of X4 virus detection at baseline. The detection limit for X4 in tropism assays is ill defined around 10%. In the current study, quantification of X4-tropic minority populations was assessed on artificial mixed samples and 38 clinical isolates. These mixtures were subjected to tropism “clonal genotyping” or “population phenotyping”. The detection of minority variants was dependant on the input of amplifiable copies. At VL > 4 log IU/ml, X4 quantification was deemed reliable. PCR founder effect and clonal resampling might result in misrepresentation of the minority species concentration at VL < 4 log. Fourteen of the clinical isolates contained dual/mixed X4-tropic virus, 5 of which were below 10% of the virus population. Currently, there is no indication what level of X4 would lead to treatment failure. Assays aiming for the detection of minority species should express results in function of VL.
Article Information
Identifiers and Pagination:
Year: 2008Volume: 2
First Page: 8
Last Page: 14
Publisher Id: TOVJ-2-8
DOI: 10.2174/1874357900802010008
Article History:
Received Date: 11/2/2008Revision Received Date: 21/2/2008
Acceptance Date: 25/2/2008
Electronic publication date: 18/3/2008
Collection year: 2008
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.5/) which permits unrestrictive use, distribution, and reproduction in any medium, provided the original work is properly cited.
* Address correspondence to this author at Virco BVBA, Generaal De Wittelaan L11B4, 2800 Mechelen, Belgium; Tel: +32 15 461 486; Fax: +32 15 285 634; E-mail: ivanden3@vrcbe.jnj.com§ Contributed equally to this work.
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 11-2-2008 |
Original Manuscript | Minor Variant Detection at Different Template Concentrations in HIV-1 Phenotypic and Genotypic Tropism Testing | |
INTRODUCTION
Recently, the FDA has approved maraviroc (Selzentry), the first of a new class of anti-Human Immunodeficiency virus 1 (HIV-1) therapeutics that targets HIV by blocking its entry into cells [1FDA. News: FDA Approves Novel Antiretroviral Drug [press release] 2007.]. Maraviroc was approved for use in combination with other antiretroviral drugs in HIV-1 experienced adult patients infected with only CCR5-tropic virus. The approval of maraviroc is based on safety and effectiveness data from 24-week data from two double-blind, placebo-controlled studies (MOTIVATE 1 and 2) with over 1000 clinical trial participants [2Lalezari J, Goodrich J, DeJesus E, et al. Efficacy and Safety of Maraviroc plus Optimized Background Therapy in Viremic, ART-experienced Patients Infected with CCR5-tropic HIV-1: 24-Week Results of a Phase 2b/3 Study in the US and Canada. 14th Conference on Retroviruses and Opportunistic Infections; 2007 February 25-28; Los Angeles California. 2007., 3Nelson M, Fätkenheuer G, Konourina I, et al. Efficacy and Safety of Maraviroc plus Optimized Background Therapy in Viremic, ART-experienced Patients Infected with CCR5-tropic HIV-1 in Europe, Australia, and North America: 24-Week Results. 14th Conference on Retroviruses and Opportunistic Infections; 2007 February 25-28; Los Angeles California. 2007.]. The label indicates that tropism testing and treatment history should guide the use of maraviroc, and that the use of maraviroc is not recommended in patients with dual/mixed or CXCR4-tropic HIV-1 [4FDA. Maraviroc Prescribing Information 2007; 23.].
A phenotypic tropism test (Trofile, Monogram Biosciences, CA; [5Whitcomb JM, Huang W, Fransen S, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism Antimicrob Agents Chemother 2007; 51(2): 566-75.]) was used during the MOTIVATE clinical trials to identify patients appropriate for treatment with maraviroc. The sensitivity of the Trofile test was determined after mixing of molecular clones, not viruses, which revealed a 100% sensitivity in picking up 10% X4-variants and 85% sensitivity at the 5% level [5Whitcomb JM, Huang W, Fransen S, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism Antimicrob Agents Chemother 2007; 51(2): 566-75.]. A total of 50 to 60% of patients with only R5 viruses were enrolled [2Lalezari J, Goodrich J, DeJesus E, et al. Efficacy and Safety of Maraviroc plus Optimized Background Therapy in Viremic, ART-experienced Patients Infected with CCR5-tropic HIV-1: 24-Week Results of a Phase 2b/3 Study in the US and Canada. 14th Conference on Retroviruses and Opportunistic Infections; 2007 February 25-28; Los Angeles California. 2007., 3Nelson M, Fätkenheuer G, Konourina I, et al. Efficacy and Safety of Maraviroc plus Optimized Background Therapy in Viremic, ART-experienced Patients Infected with CCR5-tropic HIV-1 in Europe, Australia, and North America: 24-Week Results. 14th Conference on Retroviruses and Opportunistic Infections; 2007 February 25-28; Los Angeles California. 2007.]. Treatment failure on maraviroc was associated with detection of CXCR4-tropic or dual/mixed tropic virus that was not detected by the tropism assay prior to treatment [4FDA. Maraviroc Prescribing Information 2007; 23., 6Lewis M, Simpson P, Fransen S, et al. CXCR4-using Virus Detected in Patients Receiving Maraviroc in the Phase 3 studies MOTIVATE 1 and 2 Originates from a Pre-existing Minority of CXCR4-using Virus. 16th International HIV Drug Resistance Workshop; 2007 June 12-16; Barbados. 2007.]. As the label for maraviroc indicates that prescriptions and use should be guided by tropism testing and treatment history, and for treatment of CCR5-tropic virus only, the presence of X4-tropic virus as detected by assays with cut-offs below 10% brings a new challenge to the treatment and prescription paradigm.
We developed a tropism testing platform including a genotypic and phenotypic procedure at a population and clonal level [7Van Baelen K, Vandenbroucke I, Rondelez E, Van Eygen V, Vermeiren H, Stuyver LJ. HIV-1 coreceptor usage determination in clinical isolates using clonal and population-based genotypic and phenotypic assays J Virol Methods 2007; 146(1-2): 61-73.]. The genotypic and phenotypic assays accept amplicons spanning the aminoterminal through the V4-loop (NH2-V4) of the HIV-1 Envelope gene. The presence of X4-tropic virus in the quasispecies in several clinical isolates of therapy-naïve individuals was previously illustrated [7Van Baelen K, Vandenbroucke I, Rondelez E, Van Eygen V, Vermeiren H, Stuyver LJ. HIV-1 coreceptor usage determination in clinical isolates using clonal and population-based genotypic and phenotypic assays J Virol Methods 2007; 146(1-2): 61-73.].
In the experiments described inhere, we i) further substantiated the presence of X4-tropic virus at low percentages (below 10%) in a random selection of clinical isolates, and ii) analyzed simulations of mixed populations at different viral loads.
MATERIALS AND METHODOLOGY
Sample Selection, and Tropism Determination
Properly consented plasma samples (including naïve and therapy-experienced) were randomly selected and analyzed as described in detail in Van Baelen et al. [7Van Baelen K, Vandenbroucke I, Rondelez E, Van Eygen V, Vermeiren H, Stuyver LJ. HIV-1 coreceptor usage determination in clinical isolates using clonal and population-based genotypic and phenotypic assays J Virol Methods 2007; 146(1-2): 61-73.]. In the genotypic assay: from 29 (out of 38) clinical isolates a total of 95 individual clones were analyzed, and an average of 80 ± 7 (range 62-88, Table 2) clones gave quality approved sequences. For the remaining 9 (out of 38) samples, 47 recombinant clones were analyzed, resulting in 40 ± 7 (range 23 – 47, Table 2) sequences useful for tropism prediction using PSSM
(http://ubik.microbiol.washington.edu/computing/pssm/) and SVM (http://co-receptor.bioinf.mpi-sb.mpg.de/cgi-bin/co-receptor.pl).
Preparation of Plasmids and Recombinant Virus Stocks (RVS)
The gp120 region from pNL4.3 (X4-tropic; Genbank Acc. No. AF324493) or pYK-JRCSF (R5-tropic; Genbank Acc. No. M38429) was amplified and cloned into pHXB2D-ΔNH2-V4-eGFP as described [7Van Baelen K, Vandenbroucke I, Rondelez E, Van Eygen V, Vermeiren H, Stuyver LJ. HIV-1 coreceptor usage determination in clinical isolates using clonal and population-based genotypic and phenotypic assays J Virol Methods 2007; 146(1-2): 61-73.]. Recombinant virus stocks (RVS) were generated by plasmid nucleofection (Amaxa Biosystems, Cologne, Germany) as described [7Van Baelen K, Vandenbroucke I, Rondelez E, Van Eygen V, Vermeiren H, Stuyver LJ. HIV-1 coreceptor usage determination in clinical isolates using clonal and population-based genotypic and phenotypic assays J Virol Methods 2007; 146(1-2): 61-73.] except that precautions were taken to remove contaminating plasmid DNA by washing the cells 16 hours post nucleofection. The RVS (referred to as recNL4.3 and recJRCSF) were harvested 48 hours post-infection.
Quantification of Viral RNA in RVS
Viral load (VL) of recJRCSF and recNL4.3 RVS was determined on a 1/10 dilution series using the EasyMAG - Nuclisens EasyQ HIV-1 method (Biomérieux, Boxtel, The Netherlands). Viral load values obtained were averaged and revealed that the recJRCSF RVS and recNL4.3 RVS contained 8.7 +/- 0.16 log IU/ml and 9.4 +/- 0.13 log IU/ml, respectively.
Quantification of Plasmid DNA in RVS
The presence of plasmid DNA, relative to the total nucleic acid content, was measured using real-time (RT-)PCR. RNA was quantified using primer HIV-1-F (5’-TGGGTTAT GAACTCCATCCTGAT-3’), HIV-1-R2 (5’-TGTCATTGA CAGTCCAGCTGT-3’) and probe (6-FAM-TTTCTGGCA GCACTATAGGCTGTACTGTCCATT-TAMRA). Similarly, DNA was quantified in experiments without reverse transcriptase added. recJRCSF and recNL4.3 RVS RNA isolates contained 0.31 % and 0.39 % copies plasmid, respectively.
Preparation of Mixtures of RVS, and RNA Extraction
Appropriate amounts of quantified recJRCSF and recNL4.3 RVS were used to reconstitute a set of viral mixes to yield final recNL4.3 fractions of 2.5% and 1% at 5 log IU/ml, 20% and 8% at 4 log IU/ml, and 20% at 3 log IU/ml. RNA extraction was performed on 3 to 6 aliquots of 300 µl each (EasyMAG).
Restriction Analysis
AluI restriction digestion was performed on the colony PCR products in order to distinguish between clones from recNL4.3 (4 fragments) or recJRCSF (3 fragments).
RESULTS
Tropism Analysis of Reconstituted Samples According to Viral Load
Since the exact composition of the viral quasispecies in clinical isolates is unknown, we aimed to mimic minority species conditions on artificial lab-generated virus mixtures.
The theoretical number of recNL4.3 copies, present in each RT-PCR reaction was calculated for different viral loads, and plotted as a function of the fraction recNL4.3 in the original viral mixture (Fig. 1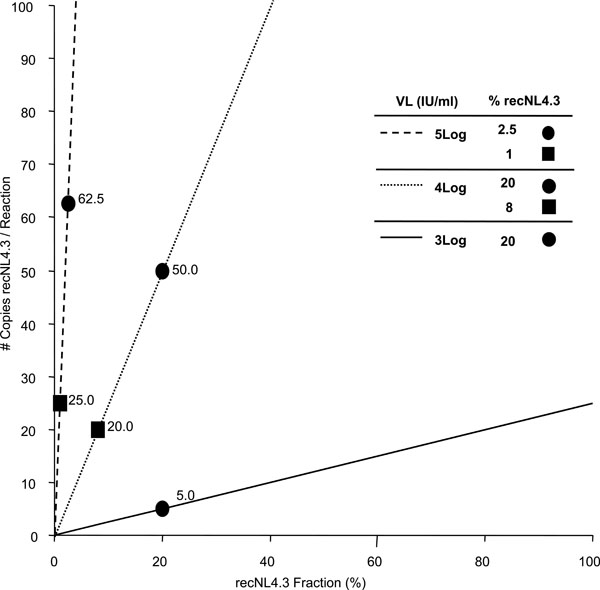 ). From this Figure one can deduce that, if, for example, the extraction/ amplification procedure starts with 300 μl plasma, eluted in 60 μl H2O, of which 5 μl is used in RT-PCR, the 20% minority fraction at 3 log IU/ml corresponds to only 5 copies. Hence, the ability to detect minority quasispecies depends on initial viral load and assay design (amount of RNA copies used for RT-PCR).
). From this Figure one can deduce that, if, for example, the extraction/ amplification procedure starts with 300 μl plasma, eluted in 60 μl H2O, of which 5 μl is used in RT-PCR, the 20% minority fraction at 3 log IU/ml corresponds to only 5 copies. Hence, the ability to detect minority quasispecies depends on initial viral load and assay design (amount of RNA copies used for RT-PCR).
 |
Fig. (2) Workflow for genotypic and phenotypic analysis of the recJRCSF/recNL4.3 mixtures. Two RVS were mixed at different ratios and viral loads. Each mixture was split into 6 aliquots and processed in parallel. RNA was extracted of each aliquot, followed by 7 replicate amplifications of the NH2-V4 region. PCR products were pooled, purified and cloned. Colonies were picked, the insert was PCR amplified, purified and analyzed by AluI restriction analysis. Tropism was assessed by population-based phenotyping as described [7Van Baelen K, Vandenbroucke I, Rondelez E, Van Eygen V, Vermeiren H, Stuyver LJ. HIV-1 coreceptor usage determination in clinical isolates using clonal and population-based genotypic and phenotypic assays J Virol Methods 2007; 146(1-2): 61-73.]. Infection of CCR5- and CXCR4-expressing U87-cells was scored using a fluorescent microscope and FACS. |
Subsequently, artificial viral mixtures panels were analyzed as visualized in Fig. (2 ), and results are shown in Table 1. At 5 log IU/ml, a 1% minority variant was detected in one out of three repeat experiments, while a 2.5% minority variant was detected in four out of six repeat experiments. At 4 log IU/ml, the minority detection success rate was 6 out of 6 (100%) for an 8% and 20% minority population. At 3 log IU/ml, the 20% minority variant was detected in three out of five experiments. At 5 and 4 log IU/ml, fluctuations between the different outcomes of the parallel experiments were relatively low, while at 3 log IU/ml, detected quantities were very different and varied from 0 to 47.3%. This can be ascribed to the initial founder effect in PCR reactions with low copy numbers, and visualized/enhanced by resampling during colony picking. When analyzing n random amplification products from an RT-PCR performed with a sample containing D RNA molecules, the expected number of independent clones (E) is given by E = D(1-((D-1)/D)n) [8Airaksinen A, Pariente N, Menendez-Arias L, Domingo E. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis Virology 2003; 311(2): 339-49., 9Liu SL, Rodrigo AG, Shankarappa R, et al. HIV quasispecies and resampling Science 1996; 273(5274): 415-6.]. For example at 3 log IU/ml, 175 RNA copies (= D) were inserted into the RT-PCR and 93 clones were picked (= n), hence E = 175(1-((175-1)/175)93) = 72. Applied to our experimental conditions, resampling might occur at a viral load of approximately 4 log or less. A selection of the samples was also tested in the population-based phenotypic analysis. At 5 log IU/ml, a 1% X4-tropic minority species was detected (Table 1). In the 2.5% recNL4.3 experiment in which no recNL4.3 could be detected genotypically, there was also no X4-tropic virus detected by phenotypically. At 4 log IU/ml, the minority species were detected in all experiments; at 3 logIU/ml, the minority was detected in two out of three experiments.
), and results are shown in Table 1. At 5 log IU/ml, a 1% minority variant was detected in one out of three repeat experiments, while a 2.5% minority variant was detected in four out of six repeat experiments. At 4 log IU/ml, the minority detection success rate was 6 out of 6 (100%) for an 8% and 20% minority population. At 3 log IU/ml, the 20% minority variant was detected in three out of five experiments. At 5 and 4 log IU/ml, fluctuations between the different outcomes of the parallel experiments were relatively low, while at 3 log IU/ml, detected quantities were very different and varied from 0 to 47.3%. This can be ascribed to the initial founder effect in PCR reactions with low copy numbers, and visualized/enhanced by resampling during colony picking. When analyzing n random amplification products from an RT-PCR performed with a sample containing D RNA molecules, the expected number of independent clones (E) is given by E = D(1-((D-1)/D)n) [8Airaksinen A, Pariente N, Menendez-Arias L, Domingo E. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis Virology 2003; 311(2): 339-49., 9Liu SL, Rodrigo AG, Shankarappa R, et al. HIV quasispecies and resampling Science 1996; 273(5274): 415-6.]. For example at 3 log IU/ml, 175 RNA copies (= D) were inserted into the RT-PCR and 93 clones were picked (= n), hence E = 175(1-((175-1)/175)93) = 72. Applied to our experimental conditions, resampling might occur at a viral load of approximately 4 log or less. A selection of the samples was also tested in the population-based phenotypic analysis. At 5 log IU/ml, a 1% X4-tropic minority species was detected (Table 1). In the 2.5% recNL4.3 experiment in which no recNL4.3 could be detected genotypically, there was also no X4-tropic virus detected by phenotypically. At 4 log IU/ml, the minority species were detected in all experiments; at 3 logIU/ml, the minority was detected in two out of three experiments.
From these experiments the following conclusions were made: i) qualitative detection of the minority species of the total population is possible genotypically and phenotypically between 3 to 5 log IU/ml; ii) quantification of the minority species is possible genotypically, but only if the viral load is higher than 4 log IU/ml, and it requires multiple separate amplifications and a high amount of colonies to analyze (Table 1); iii) quantification of the minority species in samples with a viral load below 4 log IU/ml is technically challenging and it might contain the bias of founder effect amplification (PCR drift).
Tropism Analysis of Clinical Samples
With the availability of genotyping and phenotyping protocols – including a minority species quantification if viral load is > 4 log of IU/ml in the genotypic assay - a total of 38 random clinical samples were analyzed (Table 2). In 39% of the samples (15/38), both algorithms predicted the presence of X4, of which 7 (47%) had X4 frequencies below 10%, and 4 samples (27%) below 5%. Five out of the 7 samples with X4 virus below 10% (samples 25, 26, 27, 32 and 33) were resulting in the population–based phenotyping as dual/mixed tropic (D/M) virus. Two samples that contained a subpopulation of X4 predicted clones at ± 2.5% (samples 1 and 2) resulted in the phenotypic assay as R5-tropic only. From another 19 samples, clonal sequences with discordant predictions were obtained, but all of these belonged to samples that were phenotypically R5-only. One sample contained D/M virus (sample 37), which could not be detected using genotypic analysis.
The formula E = D(1-((D-1)/D)n) [8Airaksinen A, Pariente N, Menendez-Arias L, Domingo E. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis Virology 2003; 311(2): 339-49., 9Liu SL, Rodrigo AG, Shankarappa R, et al. HIV quasispecies and resampling Science 1996; 273(5274): 415-6.] was also applied to these experiments (Table 2), again illustrating that resampling occurred at a viral load of approximately 4 log or less. Especially in the D/M samples with low copy number input, PCR founder effects and resampling could bias the results and might lead to high percentages of X4 clonal sequences. Also, the detection of the minor percentages of X4 tropic virus was only in samples with a viral load higher than 4 log IU/ml.
The conclusions of these experiments were as follows: i) X4 tropic virus is often present in clinical isolates at low percentages (<10% of the total population) and might remain undetected in some routine diagnostic tests; ii) below 4 log IU/ml, genotypic results are qualitative; iii) the qualitative phenotypic assay applied in these experiments detects most of the X4 predicted genotypes; and iv) several hundreds of genotypic clones were found discordant in the prediction algorithms (as was also recently described [10Low AJ, Dong W, Chan D, et al. Current V3 genotyping algorithms are inadequate for predicting X4 co-receptor usage in clinical isolates AIDA 2007; 21(14): F17-24.]).
DISCUSSION
CCR5 antagonists belong to the co-receptor antagonist drug class. Maraviroc, also known as Selzentry or Celsentri, is the first to receive accelerated approval by the FDA for use in combination therapy in adults infected with only CCR5-tropic HIV-1. Clinical resistance to maraviroc has not yet been fully defined, but virologic failure has been associated with the unmasking of X4-tropic virus by reducing the R5-tropic majority population, currently also interpreted as viral tropism switches [4FDA. Maraviroc Prescribing Information 2007; 23.]. The FDA states that screening for X4 viruses by HIV-1 tropism testing prior to therapy with a coreceptor antagonist should guide the use of maraviroc [4FDA. Maraviroc Prescribing Information 2007; 23.]. Several assays are available (commercially and non-commercially) that determine coreceptor usage. Some of these are based on genotypic analysis of (parts of) the envelope gene, which are either sequenced and interpreted with an algorithm [7Van Baelen K, Vandenbroucke I, Rondelez E, Van Eygen V, Vermeiren H, Stuyver LJ. HIV-1 coreceptor usage determination in clinical isolates using clonal and population-based genotypic and phenotypic assays J Virol Methods 2007; 146(1-2): 61-73., 11Sierra S, Kaiser R, Thielen A, Lengauer T. Genotypic coreceptor analysis Eur J Med Res 2007; 12(9): 453-62.], or tropism detection is based on differential hybridization of an amplicon to a probe set [12Hamy F, Vidal V, Hubert S, Klimkait T. Hybridization-based assay and replicative phenotyping as diagnostic platform for determination of co-receptor tropism. 5th European HIV Drug Resistance Workshop; 2007 March 28-30; Cascais, Portugal. 2007., 13Nelson JA, Fiscus SA, Swanstrom R. Evolutionary variants of the human immunodeficiency virus type 1 V3 region characterized by using a heteroduplex tracking assay J Virol 1997; 71(11): 8750-.]. Genotypic assays are generally fast and cheap. They rely however on the current knowledge about mutations and polymorphisms associated with co-receptor usage (interpretation algorithms or reference DNA probes). As the correlative databases will increase and algorithms will be improved, these methods might gain in importance. Phenotypic methods are based on viral entry into host cells either containing the CCR5 or CXCR4 co-receptor. Recombinant viruses are prepared by cloning or recombining (part of) the envelope gene into an HIV-1 backbone deleted for the corresponding amplicon. They consist either of a single-cycle assay (Trofile and Phenoscript [5Whitcomb JM, Huang W, Fransen S, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism Antimicrob Agents Chemother 2007; 51(2): 566-75., 14Labrosse B, Labernardiere JL, Dam E, et al. Baseline susceptibility of primary human immunodeficiency virus type 1 to entry inhibitors J Virol 2003; 77(2): 1610-3., 15Trouplin V, Salvatori F, Cappello F, et al. Determination of coreceptor usage of human immunodeficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay J Virol 2001; 75(1): 251-9.] or multiple cycle assay (PhenXR and Antivirogram [7Van Baelen K, Vandenbroucke I, Rondelez E, Van Eygen V, Vermeiren H, Stuyver LJ. HIV-1 coreceptor usage determination in clinical isolates using clonal and population-based genotypic and phenotypic assays J Virol Methods 2007; 146(1-2): 61-73., 12Hamy F, Vidal V, Hubert S, Klimkait T. Hybridization-based assay and replicative phenotyping as diagnostic platform for determination of co-receptor tropism. 5th European HIV Drug Resistance Workshop; 2007 March 28-30; Cascais, Portugal. 2007.]). The limit of detection for X4-virus in the single cycle assays is around 5-10% [5Whitcomb JM, Huang W, Fransen S, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism Antimicrob Agents Chemother 2007; 51(2): 566-75., 16Roulet V, Rochas S, Labernardière J, et al. HIV-1 PHENOSCRIPT ENV: A Sensitive Assay for the Detection of HIV X4 Minority Species and Determination of Non-B Subtype Viral Tropism. 14th Conference on Retroviruses and Opportunistic Infections; 2007 February 25-28; Los Angeles, California. 2007.]. It is not clear what level of X4-tropic virus would lead to treatment failure and what degree of X4 increase is reason for concern.
Sensitivity of an assay towards detection of a minor population is assessed in some studies by analyzing mixtures of amplicons [17Labernardière J, Lebel-binay S, Faudon JL, Holguin A, Soriano V, Chéret A. Tropism determination and performance of Phenoscript HIV-1 entry inhibitors assay Antiviral Therapy 2004; 9: S141.] or plasmids [5Whitcomb JM, Huang W, Fransen S, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism Antimicrob Agents Chemother 2007; 51(2): 566-75., 18Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase J Clin Microbiol 1999; 37(7): 2291-6.]. Extraction and amplification of viral RNA was not included in such evaluations, although these steps are an integral part of the assay workflow and contribute crucially and significantly to its sensitivity. We mimicked clinical isolates by mixing an X4-tropic minority variant with R5 tropic virus at different viral loads. The proportion of minority variants that can be detected is inversely proportional to the viral load of the sample, as theoretically anticipated [19Hughes JP, Totten P. Estimating the accuracy of polymerase chain reaction-based tests using endpoint dilution Biometrics 2003; 59(3): 505-11.]. It might be unrealistic to expect detection of minority variants below 1% at 5 log IU/ml, with decreasing chance of success at even lower viral load.
At 3 log IU/ml, and a 20% minority component, expected vs measured outcomes differed severely. This bias in template-to-product ratio is known as PCR drift and is caused by stochastic variation in the early cycles of the reaction, leading to a deviating estimation of the template frequency, especially at low copy numbers [20Becker-Pergola G, Mellquist JL, Eshleman JR, Brooks Jackson J, Eshleman SH. Improved detection of human immunodeficiency virus type 1 variants by analysis of replicate amplification reactions: Relevance to studies of human immunodeficiency virus type 1 vertical transmission Mol Diagn 1999; 4(4): 261-8., 21Polz MF, Cavanaugh CM. Bias in template-to-product ratios in multitemplate PCR Appl Environ Microbiol 1998; 64(10): 3724-0.]. In addition, the calculated number of copies present in the sample might be substantially lower than measured by the VL assay, which is based on amplification of a very small fragment (± 80 bp). In contrast, the amplification of the longer NH2-V4 fragment (± 1,3 Kb) is much more dependent on the presence of intact RNA.
A second problem at low viral load might be the issue of resampling [9Liu SL, Rodrigo AG, Shankarappa R, et al. HIV quasispecies and resampling Science 1996; 273(5274): 415-6.]. Resampling occurs if one or only a few molecules of the sample are used as input for the PCR and results in repeated analysis of PCR products amplified from the same amplifiable viral RNA molecule. This can result in artificially low levels of viral diversity, not representative for the original sample. To avoid resampling, the maximal amount of analyzed clones should not exceed the number of inserted copies into the RT-PCR [8Airaksinen A, Pariente N, Menendez-Arias L, Domingo E. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis Virology 2003; 311(2): 339-49., 9Liu SL, Rodrigo AG, Shankarappa R, et al. HIV quasispecies and resampling Science 1996; 273(5274): 415-6.].
There is growing evidence that minor X4 subpopulations are common among HIV-1 patients [22Kapoor A, Jones M, Shafer RW, Rhee SY, Kazanjian P, Delwart EL. Sequencing-based detection of low-frequency human immunodeficiency virus type 1 drug-resistant mutants by an RNA/DNA heteroduplex generator-tracking assay J Virol 2004; 78(13): 7112-23., 23Palmer S, Kearney M, Maldarelli F, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis J Clin Microbiol 2005; 43(1): 406-13.]. As viral tropism determination assays will become more mature and more sensitive, X4-tropic virus will be detected in many more patients samples. It needs to be determined what the clinical significance of these populations is and what the implications are for co-receptor antagonist use. Our results show the quantification limitations in PCR technologies on samples with a low amount of amplifiable genomes. This phenomenon is inherent to all PCR-based assays (including testing for protease and RT gene resistance mutations) on small amounts of complex templates [24Karrer EE, Lincoln JE, Hogenhout S, et al. In situ isolation of mRNA from individual plant cells: creation of cell-specific cDNA libraries Proc Natl Acad Sci USA 1995; 92(9): 3814-8.] and should be carefully considered when samples are analyzed with low input copy number. Assays aiming for minority species detection are limited by the amount of copies inserted into the RT-PCR procedure and should express results in function of the number of target molecules present in the original sample.
ACKNOWLEDGEMENTS
We would like to thank Bart Winters for helpful discussions, and Lee Bacheler and Jorge Villacian for critical reading of the manuscript.





