- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Influenza A(H1N1) Oseltamivir Resistant Viruses in the Netherlands During the Winter 2007/2008
Frederika Dijkstra*, 1, Marcel Jonges 1, 2, Ruud van Beek 2, Gé A Donker 3, François G Schellevis 3, 4, Marion Koopmans 1, 2, Marianne A.B van der Sande 1, 5, Albert D.M.E Osterhaus 2, Charles A.B Boucher 2, Guus F Rimmelzwaan 2, Adam Meijer 1
Abstract
Background:
Antiviral susceptibility surveillance in the Netherlands was intensified after the first reports about the emergence of influenza A(H1N1) oseltamivir resistant viruses in Norway in January, 2008.
Methods:
Within the existing influenza surveillance an additional questionnaire study was performed to retrospectively assess possible risk factors and establish clinical outcome of all patients with influenza virus A(H1N1) positive specimens. To discriminate resistant and sensitive viruses, fifty percent inhibitory concentrations for the neuramidase inhibitors oseltamivir and zanamivir were determined in a neuraminidase inhibition assay. Mutations previously associated with resistance to neuramidase inhibitors and M2 blockers (amantadine and rimantadine) were searched for by nucleotide sequencing of neuraminidase and M2 genes respectively.
Results:
Among 171 patients infected with A(H1N1) viruses an overall prevalence of oseltamivir resistance of 27% (95% CI: 20-34%) was found. None of influenza A(H1N1) oseltamivir resistant viruses tested was resistant against amantadine or zanamivir. Patient characteristics, underlying conditions, influenza vaccination, symptoms, complications, and exposure to oseltamivir and other antivirals did not differ significantly between patients infected with resistant and sensitive A(H1N1) viruses.
Conclusion:
In 2007/2008 a large proportion of influenza A(H1N1) viruses resistant to oseltamivir was detected. There were no clinical differences between patients infected with resistant and sensitive A(H1N1) viruses. Continuous monitoring of the antiviral drug sensitivity profile of influenza viruses is justified, preferably using the existing sentinel surveillance, however, complemented with data from the more severe end of the clinical spectrum. In order to act timely on emergencies of public health importance we suggest setting up a surveillance system that can guarantee rapid access to the latter.
Article Information
Identifiers and Pagination:
Year: 2011Volume: 5
First Page: 154
Last Page: 162
Publisher Id: TOVJ-5-154
DOI: 10.2174/1874357901105010154
Article History:
Received Date: 30/8/2011Revision Received Date: 27/10/2011
Acceptance Date: 28/10/2011
Electronic publication date: 23/12/2011
Collection year: 2011
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http: //creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Centre for Infectious Disease Control, National Institute for Public Health and the Environment, PO box 1 – pb 75, 3720 BA Bilthoven, The Netherlands; Tel: +31 30 2743781; Fax: +31 30 2744409; E-mail: frederika.dijkstra@rivm.nl
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 30-8-2011 |
Original Manuscript | Influenza A(H1N1) Oseltamivir Resistant Viruses in the Netherlands During the Winter 2007/2008 | |
INTRODUCTION
Infections with influenza viruses are responsible for substantial morbidity and mortality each year during the winter months [1Simonsen L. The global impact of influenza on morbidity and mortality Vaccine 1999; 17(Suppl 1): S3-10.]. Although vaccination is the main method of preventing influenza associated illness and death, antivirals may offer a valuable addition for influenza patients at high risk for complications and patient groups that cannot be vaccinated effectively. The influenza virus neuraminidase inhibitors (NAI) zanamivir and oseltamivir were introduced in 1999. Similar to most other European countries, antivirals are not widely used in the Netherlands to treat seasonal influenza and to prevent complications [2Kramarz P, Monnet D, Nicoll A, Yilmaz C, Ciancio B. Use of oseltamivir in 12 European countries between 2002 and 2007--lack of association with the appearance of oseltamivir-resistant influenza A(H1N1) viruses Eur Surveill 2009; 14(ii): 19112.]. Antivirals are mostly used therapeutically and prophylactically during influenza outbreaks in nursing homes and for the treatment of infected immunocompromised patients in hospitals. In addition, they are stockpiled as part of pandemic preparedness plans [3Meijer A, Lackenby A, Hay A, Zambon M. Influenza antiviral susceptibility monitoring activities in relation to national antiviral stockpiles in Europe during the winter 2006/2007 season Eur Surveill 2007; 12: E3-4.].
Nevertheless, monitoring of influenza virus antiviral susceptibility has become more important [3Meijer A, Lackenby A, Hay A, Zambon M. Influenza antiviral susceptibility monitoring activities in relation to national antiviral stockpiles in Europe during the winter 2006/2007 season Eur Surveill 2007; 12: E3-4.]. In the Netherlands, the National Institute for Public Health and the Environment (RIVM) monitored susceptibility of influenza viruses to antiviral drugs within the network of sentinel general practitioners since the 2005/2006 winter season [4Jonges M, Van Der Lubben IM, Dijkstra F, Verhoef L, Koopmans M, Meijer A. Dynamics of antiviral-resistant influenza viruses in the Netherlands, 2005-2008 Antiviral Res 2009; 83: 290-7.].
In 2005/2006 and 2006/2007 in the Netherlands, viruses with a lower sensitivity to the NAI were found only sporadically (2%), whilst a high proportion of A(H3N2) viruses were found to be resistant to the M2 ion-channel inhibitors (M2I) amantadine and rimantadine [4Jonges M, Van Der Lubben IM, Dijkstra F, Verhoef L, Koopmans M, Meijer A. Dynamics of antiviral-resistant influenza viruses in the Netherlands, 2005-2008 Antiviral Res 2009; 83: 290-7.]. Until 2007, NAI resistant influenza viruses were also rarely observed elsewhere in the world [5Escuret V, Frobert E, Bouscambert-Duchamp M, et al. Detection of human influenza A (H1N1) and B strains with reduced sensitivity to neuraminidase inhibitors J Clin Virol 2008; 41: 25-8.-8Mungall BA, Xu X, Klimov A. Surveillance of influenza isolates for susceptibility to neuraminidase inhibitors during the 2000-2002 influenza seasons Virus Res 2004; 103: 195-7.]. However, by the end of January 2008, the emergence of influenza A(H1N1) oseltamivir resistant viruses (ORV) was first reported in Norway [9ECDC. Timeline for the emergence of oseltamivir resistant influenza A(H1N1) 2007-8 Available from: http://ecdc.europa.eu/en/files/pdf/Health_topics/ECDC_AVR_timeline_v1_31-10-08.pdf [cited 2008 November 21];, 10Hauge SH, Dudman S, Borgen K, Lackenby A, Hungnes O. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007-08 Emerg Infect Dis 2009; 15: 155-62.], and shortly thereafter in several other European countries [11Lackenby A, Hungnes O, Dudman SG, et al. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe Eur Surveill 2008; 13(ii): 8026.]. The resistance was associated with a amino acid substitution at position 275 (H275Y or H274Y in N2 numbering) of the neuraminidase, which confers a high level of resistance against oseltamivir [11Lackenby A, Hungnes O, Dudman SG, et al. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe Eur Surveill 2008; 13(ii): 8026.].
Clinical trials showed that resistance against oseltamivir generally developed at low incidence in patients treated with oseltamivir (0.32% in adults and 4.1% in children) [12Aoki FY, Boivin G, Roberts N. Influenza virus susceptibility and resistance to oseltamivir Antivir Ther 2007; 12: 603-16.]. However, in two small Japanese studies oseltamivir-resistance emerged in 18% (9/50) and 16% (7/43) of treated Japanese children with influenza virus A(H3N2) and A(H1N1) infection respectively [13Kiso M, Mitamura K, Sakai-Tagawa Y, et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study Lancet 2004; 364: 759-65., 14Ward P, Small I, Smith J, Suter P, Dutkowski R. Oseltamivir (Tamiflu) and its potential for use in the event of an influenza pandemic J Antimicrob Chemother 2005; 55(Suppl 1): i5-i21.]. Nevertheless, the overall evidence before the 2007/2008 season suggested that influenza A(H1N1) viruses carrying the H275Y substitution were highly compromised: their replicative potential was reduced and transmissibility and pathogenicity was decreased [12Aoki FY, Boivin G, Roberts N. Influenza virus susceptibility and resistance to oseltamivir Antivir Ther 2007; 12: 603-16., 15Ives JA, Carr JA, Mendel DB, et al. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo Antiviral Res 2002; 55: 307-17.].
Immediately after the first reports about the relative high prevalence of ORV in several European countries, the antiviral susceptibility surveillance in the Netherlands was intensified and besides influenza A(H1N1) viruses obtained from sentinel patients (patients consulting a sentinel general practitioner [GP]), non-sentinel patients (mainly hospitalised patients) were included. In addition, a follow-up questionnaire study was started to determine the clinical characteristics of and possible risk factors for infection with ORV in the Netherlands. Here we report our findings of these integrated epidemiological and virological studies performed during the winter season 2007/2008.
METHODS
Influenza Surveillance
In the Netherlands, the influenza surveillance is carried out by the NIVEL (Netherlands Institute of Health Services Research) and the Dutch National Influenza Centre (NIC, a collaboration between the RIVM and the Erasmus Medical Center [ErasmusMC]). Sentinel surveillance of influenza is performed in the Dutch Sentinel Practice Network of NIVEL, a nationwide network of 45 general practices [16Donker GA. Continuous morbidity registration at Dutch sentinel stations, 2007 Utrecht: NIVEL 2008.]. The specimens from this sentinel surveillance are examined for influenza viruses by real-time reverse-transcriptase polymerase chain reaction (RT-PCR) [17Jonges M, Liu WM, van der Vries E, et al. Influenza virus inactivation for studies of antigenicity and phenotypic neuraminidase inhibitor resistance profiling J Clin Microbiol 2010; 48: 928-40.], full details available on request. Influenza virus isolates derived from PCR positive specimens in tissue culture [4Jonges M, Van Der Lubben IM, Dijkstra F, Verhoef L, Koopmans M, Meijer A. Dynamics of antiviral-resistant influenza viruses in the Netherlands, 2005-2008 Antiviral Res 2009; 83: 290-7.] are used for phenotypic determination of antiviral susceptibility and sent to NIC-ErasmusMC for antigenic characterization determining the match with the season’s vaccine virus. In addition, all diagnostic laboratories in the Netherlands are requested, on a voluntary basis, to send in their influenza virus positive specimens or virus isolates to the NIC-ErasmusMC for antigenic characterisation (non-sentinel surveillance). These specimens and viruses are mainly from hospitalized patients, because in general in the Netherlands diagnostics for influenza virus is done only in patients with a severe acute respiratory illness. Virus isolation and propagation at NIC ErasmusMC were done on Madin Darby Canine Kidney (MDCK) cells.
Antiviral Susceptibility Monitoring
All influenza A(H1N1) viruses obtained from the influenza surveillance with date of sampling between 1st October 2007 and 18th May 2008 were tested for their antiviral susceptibility. The NAI sensitivity was determined using a fluorescence based NA-inhibition assay and expressed as the concentration of NAI needed to inhibit the NA enzyme activity by 50% (IC50), as described previously [4Jonges M, Van Der Lubben IM, Dijkstra F, Verhoef L, Koopmans M, Meijer A. Dynamics of antiviral-resistant influenza viruses in the Netherlands, 2005-2008 Antiviral Res 2009; 83: 290-7., 18Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate Anal Biochem 1979; 94: 287-96., 19Wetherall NT, Trivedi T, Zeller J, et al. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors: report of the neuraminidase inhibitor susceptibility network J Clin Microbiol 2003; 41: 742-50.]. Presence or absence of the H275Y substitution in NA and amino acid substitutions associated with M2I resistance were determined by cycle-sequencing of the NA and M2 genes, respectively. For low viral load clinical specimens negative in tissue-culture, the presence or absence of H275Y was determined using pyrosequencing [11Lackenby A, Hungnes O, Dudman SG, et al. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe Eur Surveill 2008; 13(ii): 8026.]. The haemagglutinin (HA1) gene was sequenced and used to study possible evolutionary relationships between sensitive and resistant viruses. DNA sequences were assembled, edited, translated and clustered by Neighbor Joining with Jukes & Cantor correction using Bionumerics V6.5 software (Applied Maths, Sint-Martens-Latem, Belgium). Full sequencing details are available upon request.
Study on Risk Factors and Clinical Impact
To determine the possible impact of oseltamivir-resistance on the severity of disease caused by A(H1N1) influenza viruses and to identify potential risk factors for infection with ORV a study was started among all (GP and hospital) patients diagnosed with an A(H1N1) influenza virus infection during the influenza season 2007/2008. We collected baseline variables (i.e. date of birth, sex, date of onset illness, date of specimen collection), information on symptoms and diagnosis, underlying illnesses, data about the clinical course, vaccination status, and treatment with oseltamivir or other antiviral drugs. For the sentinel patients, baseline data were obtained from the clinical specimen form and for supplementary data the GPs of the patients were asked to complete an additional questionnaire which was sent by e-mail or by regular mail. For the non-sentinel patients, only date of birth, sex, date of specimen collection, and name and location of the virologist who had sent in the virus was known at NIC-ErasmusMC. Therefore, patients’ physicians were asked to complete a questionnaire to provide the baseline data as well as the follow-up data. The questionnaires were developed in collaboration with the European Influenza Surveillance Scheme, European Centre for Disease Prevention and Control, and the World Health Organization.
Data Analysis
Weekly incidence rates of ILI (influenza-like illness, defined according to the PEL criteria [16Donker GA. Continuous morbidity registration at Dutch sentinel stations, 2007 Utrecht: NIVEL 2008.]) during the winter season 2007/2008 were calculated using consultation data from sentinel GPs and were compared to weekly incidence rates of ILI during the 2000/2001 season, i.e. the previous winter season in the Netherlands in which A(H1N1) influenza viruses were dominant [20De Jong JC, Rimmelzwaan GF, Bartelds AI, Wilbrink B, Fouchier RA, Osterhaus AD. [2000/01 influenza season and the vaccine composition for the season 2001/'02] Ned Tijdschr Geneeskd 2001; 145: 1945-50., 21Dijkstra F, Donker GA, Wilbrink B, Van Gageldonk-Lafeber AB, Van Der Sande MA. Long time trends in influenza-like illness and associated determinants in the Netherlands Epidemiol Infect 2009; 137: 473-9.]. The weekly number of influenza viruses detected in the sentinel and non-sentinel surveillance was calculated per type and subtype.
Viruses with the H275Y substitution or an IC50 ≥ 100 nM for oseltamivir were considered resistant. IC50 values of the remaining viruses were used in box-and-whisker plot analysis to identify potential outliers for NAI susceptibility [22Massart DL, Smeyers-Verbeke J, Capron X, Schlesier K. Visual presentation of data by means of box plots LC-GC Europe 2005; 18: 215-18.]. IC50-values between 1.5 and 3 times the interquartile range (IQR), or more than 3 times the IQR outside the IQR were defined as mild or extreme outliers, respectively. After elimination of outliers, the remaining IC50-values were used to calculate the mean and standard deviation (SD) of baseline NAI susceptibility of Dutch influenza viruses. All IC50-values were log-transformed prior to analysis and back-transformed afterwards.
The Cochran-Armitage test for trend was used to test if there was a time trend in proportion of ORV. The Chi Square tests and Fisher exact tests were used for univariate analyses to compare possible risk factors and clinical characteristics between patients with influenza A(H1N1) ORV and oseltamivir sensitive viruses (OSV). In addition, odds ratio’s (ORs) were calculated to analyze whether potential risk factors were associated with ORV infection. In this analysis patients infected with ORV were regarded as cases and patients infected with OSV were regarded as controls (nested case-control approach). Finally, risk ratio’s (RRs) were calculated to determine the possible impact of ORV on the severity of disease. In this analysis patients infected with ORV were regarded as the exposed group and patients infected with OSV were regarded as the unexposed group (cohort approach). Analyses were done separately for sentinel and non-sentinel patients because of the major differences in for example age, symptoms, complications, and hospitalizations between these patient groups. Statistical analyses were performed using SAS software version 9.1 (SAS Institute, Cary, NC, USA).
RESULTS
ILI Incidence and Circulating Influenza Viruses in the 2007/2008 Season
During the 2007/2008 season, weekly ILI incidences were above the epidemic baseline threshold from the fifth till the tenth week of 2008, as well in weeks 12 and 13 [16Donker GA. Continuous morbidity registration at Dutch sentinel stations, 2007 Utrecht: NIVEL 2008., 23Rimmelzwaan GF, De Jong JC, Donker GA, Meijer A, Fouchier RA, Osterhaus AD. Influenza season 2007/'08 in the Netherlands: antigenic variation, oseltamivir resistance and vaccine composition for the 2008/'09 season Ned Tijdschr Geneeskd 2008; 152: 2138-44.] and peaked later than during the 2000/2001 season (Fig. 1A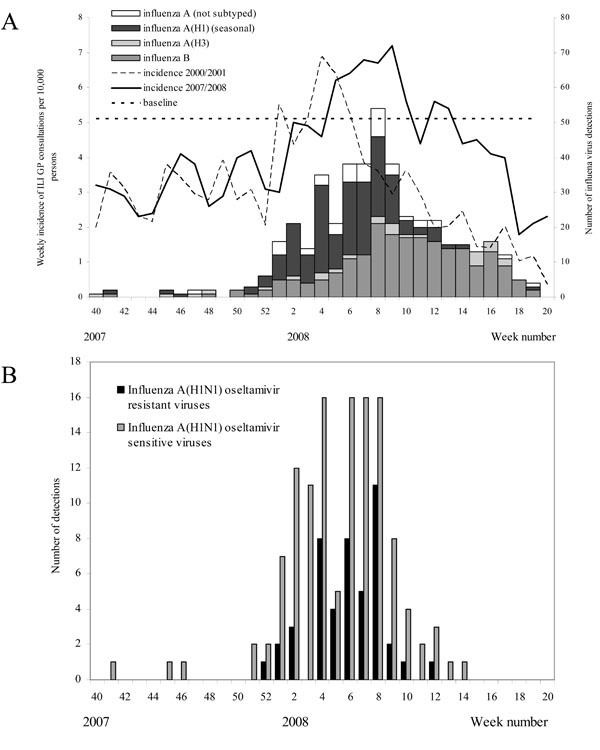 ). The mean weekly incidence of ILI in the 2007/2008 season (4.1 per 10,000 population) was somewhat higher compared to that for the 2000/2001 season (3.1 per 10,000 population), while peak incidence levels were comparable in both seasons. The number of influenza virus detections started to rise from the first week of 2008 (Fig. 1A
). The mean weekly incidence of ILI in the 2007/2008 season (4.1 per 10,000 population) was somewhat higher compared to that for the 2000/2001 season (3.1 per 10,000 population), while peak incidence levels were comparable in both seasons. The number of influenza virus detections started to rise from the first week of 2008 (Fig. 1A ). In the beginning of the epidemic most influenza viruses were of type A, while from week 10 onwards, most influenza viruses were of type B. Influenza A(H3) viruses were detected only sporadically.
). In the beginning of the epidemic most influenza viruses were of type A, while from week 10 onwards, most influenza viruses were of type B. Influenza A(H3) viruses were detected only sporadically.
Study Population
The study population consisted of patients from both sentinel and non-sentinel surveillance who tested positive for A(H1N1) influenza virus (Table 1). From the non-sentinel surveillance 100 A(H1N1) virus isolates and respiratory secretions were received during the 2007/2008 season. In the sentinel surveillance a total of 846 respiratory specimens were obtained during the 2007/2008 seasons, of which 73 (8.6%) were tested positive for A(H1N1) influenza virus by PCR. Two specimens had a too low viral load for virus isolation or pyrosequencing and therefore determination of antiviral susceptibility was not possible. Thirty-eight of the PCR-positive respiratory specimens yielded influenza A(H1N1) virus isolates, which were tested phenotypical for antiviral susceptibility and a subset of 28 by sequencing. The remaining 33 sentinel specimens were analyzed for antiviral susceptibility by pyrosequencing only. In the non-sentinel surveillance a total of 96 A(H1N1) virus isolates, which were all tested phenotypically and genotypically for antiviral susceptibility, and four A(H1N1) PCR-positive respiratory secretions, which were tested for antiviral susceptibility by H275Y detection only, were received from other laboratories during the 2007/2008 season. In total, the antiviral profile was available for viruses detected in 71 sentinel patients and 100 non-sentinel patients, which were included in the clinical follow-up study.
These specimens were sent for analysis by 30 GPs and 16 laboratories nationwide, respectively. Twenty-five GPs and physicians related to 12 laboratories responded to the questionnaires. Basic clinical data (i.e. data from clinical specimen form) were available for all 71 sentinel patients. Complete data (i.e. data from clinical specimen form and additional questionnaire) were obtained for 53/71 (74.6%) of sentinel patients. For the non-sentinel patients, complete data was obtained for 49/100 (49.0%) patients, while for 32 (32%) only sex, date of birth, geographic location, and date of sample collection was available.
Age of patients from the sentinel surveillance (median 26.2 years) was significantly higher than age of patients from the non-sentinel surveillance (median 0.9 years, p<0.001) (Table 2). Furthermore, prevalence of being immunocompromised as well as having a chronic disease was higher in patients from the non-sentinel surveillance compared to patients from the sentinel surveillance (p<0.01) (Table 2).
Sensitivity to Antiviral Drugs
Phenotypic assessment of the NAI sensitivity of all influenza A(H1N1) isolates (n=134) indicated that 29% of the viruses was resistant against oseltamivir (Table 1), but sensitive to zanamivir (mean IC50 ± SD: 0.87±0.22nM). Amino acid sequence analysis of the A(H1N1) isolates confirmed the presence of the H275Y substitution in the NA of all these ORVs. Sequence analysis of all 37 A(H1N1) virus PCR-positive specimens from which virus could not be isolated demonstrated the presence of the H275Y substitution in 7 (19%) of the samples. Sequence analysis of M2 of 46 sentinel influenza A(H1N1) viruses showed that neither in ORVs nor in OSVs known M2I resistance substitutions were detected. The combined phenotypic and genotypic data showed that 27% of Dutch influenza A(H1N1) viruses were resistant against oseltamivir (Table 1), but sensitive to zanamivir and M2I. In addition to the 39 oseltamivir resistant A(H1N1) viruses detected by phenotypic analysis, box-and-whisker plots identified three outliers with reduced zanamivir susceptibility, having IC50-values of 3.1nM, 3.4nM and 5.3nM. The baseline zanamivir IC50-value was consequently adapted from 1.05±0.61nM to 0.98±0.40nM (mean±SD).
The number of ORVs and OSVs detected by week is shown in Fig. (1B ). Oseltamivir-resistant influenza A(H1N1) viruses were collected between week 52 of 2007 until week 12 of 2008. No significant increase in the proportion ORV over time was detected (p>0.05) and no significant difference was observed in prevalence of ORV between patients from the sentinel and the non-sentinel surveillance (p>0.05).
). Oseltamivir-resistant influenza A(H1N1) viruses were collected between week 52 of 2007 until week 12 of 2008. No significant increase in the proportion ORV over time was detected (p>0.05) and no significant difference was observed in prevalence of ORV between patients from the sentinel and the non-sentinel surveillance (p>0.05).
Phylogenetic Analysis
Based on NA sequences, oseltamivir resistance of the A(H1N1) influenza viruses was solely the result of amino acid substitution H275Y, however, accompanied by amino acid substitution D354G. Within the ORV strains, two NA types could be identified, with and without silent nucleotide substitution t243g (Fig. 2 ). Based on HA1 sequences, ORV strains contained silent nucleotide substitution t162c, but also showed more amino acid variation near the receptor-binding site than OSV strains. In this region amino acid substitutions N183D, D186N/V and A189T were observed. The oseltamivir resistant NA carrying the silent t243g nucleotide substitution, was associated with amino acid substitution A189T in HA1.
). Based on HA1 sequences, ORV strains contained silent nucleotide substitution t162c, but also showed more amino acid variation near the receptor-binding site than OSV strains. In this region amino acid substitutions N183D, D186N/V and A189T were observed. The oseltamivir resistant NA carrying the silent t243g nucleotide substitution, was associated with amino acid substitution A189T in HA1.
Risk Factors and Clinical Impact
Clinical Diagnosis
Most influenza A(H1N1) infected patients from the sentinel surveillance were diagnosed with ILI (78.6% and 77.2% of the patients with ORV and OSV respectively), while patients from the non-sentinel surveillance were mostly diagnosed with Acute Respiratory Infections other than ILI (ARI) (65.0% and 82.1% of the patients with ORV and OSV respectively). Specific diagnoses for patients with other ARI from the sentinel surveillance were common cold, sinusitis, otitis media, pharyngitis, tonsillitis, and bronchitis, whereas for patients from non-sentinel surveillance these were mostly common cold, bronchiolitis, pneumonia, and other diagnoses.
Clinical Symptoms
Symptoms of patients infected with ORV and OSV are shown in Fig. (3 ). An acute start, fever, and coughing were the most frequently (>80%) reported symptoms in sentinel patients. In non-sentinel patients these symptoms were less frequently reported. No significant differences between patients with ORV and OSV were observed for any of the symptoms. In addition, no significant difference was observed in the number of reported symptoms per patient between these groups.
). An acute start, fever, and coughing were the most frequently (>80%) reported symptoms in sentinel patients. In non-sentinel patients these symptoms were less frequently reported. No significant differences between patients with ORV and OSV were observed for any of the symptoms. In addition, no significant difference was observed in the number of reported symptoms per patient between these groups.
Possible Risk Factors
Prevalence of ORV was significantly higher for sentinel patients who were sampled in a sentinel practice in the provinces North- and South-Holland (the west of the country) compared to other provinces (OR=8.6, 95% CI 2.1 – 34.9) (Table 2). None of the patients with ORV had used oseltamivir themselves in the two weeks before sampling and only one patient was exposed to a household contact who had used oseltamivir. In addition, none of the other possible risk factors were significantly associated with ORV infection (Table 2).
Clinical Course
For sentinel as well as for non-sentinel patients the course and severity of illness of infection was independent of oseltamivir sensitivity profile of the influenza A(H1N1) viruses (Table 3).
DISCUSSION
In Europe and worldwide, influenza A(H1N1) oseltamivir resistant viruses started to spread during the 2007/2008 winter season. Prevalence of resistant viruses varied considerably from country to country [6Sheu TG, Deyde VM, Okomo-Adhiambo M, et al. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008 Antimicrob Agents Chemother 2008; 52: 3284-92., 24Dharan NJ, Gubareva LV, Meyer JJ, et al. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States JAMA 2009; 301: 1034-41.-28Ciancio BC, Meerhoff TJ, Kramarz P, et al. Oseltamivir-resistant influenza A(H1N1) viruses detected in Europe during season 2007-8 had epidemiologic and clinical characteristics similar to co-circulating susceptible A(H1N1) viruses Eur Surveill 2009; 14(ii): 19412.]. In the Netherlands we found an overall prevalence of oseltamivir resistance of 26.9% (95%CI 20.3 – 33.6%) among 171 A(H1N1) viruses analysed during the winter season 2007/2008. This prevalence was somewhat higher than the average prevalence of ORV in Europe (20.1%, 95%CI 15.2-24.6%) [27Meijer A, Lackenby A, Hungnes O, et al. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 Season Emerg Infect Dis 2009; 15: 552-60.]. All ORVs in the Netherlands were still susceptible to amantadine and zanamivir.
In our study none of the patients with an ORV infection had used oseltamivir themselves in the two weeks before sampling and only one was exposed to a household contact who had used oseltamivir. Therefore, a causal relation between use of oseltamivir and ORV in the Netherlands was quite unlikely. This finding is consistent with the low number of oseltamivir prescriptions in the Netherlands (15-266 prescriptions per month in public pharmacies in the period July 2006 till June 2008 for the entire Dutch population of 16 million, source: Stichting Farmaceutische Kengetallen). Similar studies from other countries also did not demonstrate a relation between use of oseltamivir and ORV [10Hauge SH, Dudman S, Borgen K, Lackenby A, Hungnes O. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007-08 Emerg Infect Dis 2009; 15: 155-62., 24Dharan NJ, Gubareva LV, Meyer JJ, et al. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States JAMA 2009; 301: 1034-41., 28Ciancio BC, Meerhoff TJ, Kramarz P, et al. Oseltamivir-resistant influenza A(H1N1) viruses detected in Europe during season 2007-8 had epidemiologic and clinical characteristics similar to co-circulating susceptible A(H1N1) viruses Eur Surveill 2009; 14(ii): 19412.]. These findings are consistent with the lack of an association between ORV and the use of oseltamivir in 12 European countries [2Kramarz P, Monnet D, Nicoll A, Yilmaz C, Ciancio B. Use of oseltamivir in 12 European countries between 2002 and 2007--lack of association with the appearance of oseltamivir-resistant influenza A(H1N1) viruses Eur Surveill 2009; 14(ii): 19112.].
Between sentinel and non-sentinel patients there were major differences in age, hospitalization, and chronic conditions, which are related to the settings from which specimens were derived. The non-sentinel specimens represent a selection of influenza viruses from the more severe, hospitalized, patients, while sentinel specimens were from patients consulting a GP and likely present with less severe disease. Because of these differences between sentinel and non-sentinel patients, analyses were done separately for these two groups.
Both for sentinel and for non-sentinel patients age, gender, travelling abroad, underlying condition (measured as the presence or absence of a chronic disease, immunosuppression, and respiratory allergy), and vaccination status were similar for patients infected with ORV and OSV. The prevalence of ORV was significantly higher for sentinel patients that were sampled in sentinel practices in the provinces North- and South-Holland (the most densely populated provinces of the Netherlands) compared to other provinces. No clear explanation for this observation could be found.
The clinical course was similar for patients with OSR and OSV when assessing initial clinical symptoms, complications, hospitalization, and time to recovery. This finding is consistent with studies from Norway and the U.S. [10Hauge SH, Dudman S, Borgen K, Lackenby A, Hungnes O. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007-08 Emerg Infect Dis 2009; 15: 155-62., 24Dharan NJ, Gubareva LV, Meyer JJ, et al. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States JAMA 2009; 301: 1034-41.]. However, mainly milder complications could have been missed, because it is conceivable that the physicians were not always aware of the exact clinical course of the patients in the period of 4 weeks after the sampling. At the moment of the questionnaire, GPs of the sentinel surveillance did not know the oseltamivir resistance status of the patient, so for the sentinel patients this could not have affected the answers to the questions about clinical course. Physicians and virologists of the included patients from the non-sentinel surveillance could in theory be aware of the oseltamivir resistance status of their patients if testing oseltamivir resistance was also performed in a peripheral laboratory on request of the peripheral virologist and/or physicians. However, on inquiry only one of the physicians/ virologists reported that testing of oseltamivir resistance was performed in a peripheral laboratory for 3 of their patients in this study.
The relatively mild influenza season in the Netherlands, which was comparable with the 2000/2001 influenza season, suggested that on the population level, morbidity was not increased because of the emergence of ORV. However, for individuals at high risk of complications following influenza virus infection (for example immunocompromised patients), an influenza virus infection can be life-threatening in case the infection is not treated (or prevented) effectively. For these patients infection with an ORV may have had a high clinical impact and even could be fatal [29Gooskens J, Jonges M, Claas EC, Meijer A, Van Den Broek PJ, Kroes AM. Morbidity and mortality associated with nosocomial transmission of oseltamivir-resistant influenza A(H1N1) virus JAMA 2009; 301: 1042-6., 30Van Der Vries E, Van Den Berg B, Schutten M. Fatal oseltamivir-resistant influenza virus infection N Engl J Med 2008; 359: 1074-6.], although a fatal outcome could also occur following an infection with an OSV. It should be noted that our study included only one of the 3 deceased Dutch patients who were infected with an ORV and previously described in literature [29Gooskens J, Jonges M, Claas EC, Meijer A, Van Den Broek PJ, Kroes AM. Morbidity and mortality associated with nosocomial transmission of oseltamivir-resistant influenza A(H1N1) virus JAMA 2009; 301: 1042-6., 30Van Der Vries E, Van Den Berg B, Schutten M. Fatal oseltamivir-resistant influenza virus infection N Engl J Med 2008; 359: 1074-6.].
Close monitoring of HA and NA gene segments composition of resistant and sensitive viruses in conjunction with patient information offered the opportunity to analyse the impact of micro-evolution on clinical and epidemiological presentation. During the 2007-2008 influenza season, clustering of A(H1N1) virus strains was observed due to micro-evolution in HA and NA gene segments. One oseltamivir resistant A(H1N1) cluster, carrying the amino acid substitution A189T (193 in H3 numbering) near the receptor binding site of HA, was associated with the NA cluster carrying silent nucleotide substitution t243g. Influenza viruses within the A189T HA cluster were obtained from both sentinel and non-sentinel surveillance throughout the influenza season. The first was obtained from a hospitalised patient in December 2007. In recent globally detected influenza A(H3N2) viruses, variation at the same location (S193F) was associated with the fixation of adamantane resistance caused by S31N in M2 [31Saito R, Li D, Shimomura C, et al. An off-seasonal amantadine-resistant H3N2 influenza outbreak in Japan Tohoku J Exp Med 2006; 210: 21-7.]. As the A189T HA cluster proved dominant in the oseltamivir resistant A(H1N1) viruses in South Africa in 2008 [26Besselaar TG, Naidoo D, Buys A, et al. Widespread oseltamivir resistance in influenza A viruses (H1N1), South Africa Emerg Infect Dis 2008; 14: 1809-0.], we hypothesize that A189T in HA possibly re-established the HA and NA balance in recent ORV variants, as suggested by Collins et al., [32Collins PJ, Haire LF, Lin YP, et al. Structural basis for oseltamivir resistance of influenza viruses Vaccine 2009; 27: 6317-23.]. Although Norway was the first country to report unprecedented high prevalence of influenza A(H1N1) ORV at the onset of the 2007-2008 epidemic, the A189T HA cluster was not detected in Norwegian sequences [10Hauge SH, Dudman S, Borgen K, Lackenby A, Hungnes O. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007-08 Emerg Infect Dis 2009; 15: 155-62.]. This would exclude Norway as contributor for global spread and fixation of oseltamivir resistance. Moreover, as resistant viruses belonging to the A189T HA cluster simultaneously co-emerged in Canada, the United States and Europe late 2007, it suggests that acquisition of oseltamivir resistance was the result of independent introductions and was not drug induced [2Kramarz P, Monnet D, Nicoll A, Yilmaz C, Ciancio B. Use of oseltamivir in 12 European countries between 2002 and 2007--lack of association with the appearance of oseltamivir-resistant influenza A(H1N1) viruses Eur Surveill 2009; 14(ii): 19112., 27Meijer A, Lackenby A, Hungnes O, et al. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 Season Emerg Infect Dis 2009; 15: 552-60.]. Recent evidence showed that pre-existing amino acid substitutions in the neuraminidase itself (R222Q and V234M) were necessary substitutions needed for sustained transmissibility of the resistant H275Y variant [33Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance Science 2010; 328: 1272-5.]. These substitutions increase the number of NA proteins on the virus whilst the H275Y substitution reduces the number of surface expressed NA proteins, thereby balancing NA and HA activity again. Therefore, possibly in addition to the A189T substitution in HA the R222Q and V234M substitutions in NA created the prerequisites for the emergence of the H275Y variant. All the Dutch viruses of the 2007/2008 season possessed the R222Q and V234M substitutions in accordance with the data presented by Bloom et al., [33Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance Science 2010; 328: 1272-5.]. However, the D354G substitution in the NA distinguished ORV from OSV, as has been observed for all European ORV [27Meijer A, Lackenby A, Hungnes O, et al. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 Season Emerg Infect Dis 2009; 15: 552-60.]. The clinical and epidemiological studies demonstrated that no significant differences were found between symptoms and patients infected with OSV or ORV, and therefore mutations accompanying H275Y were not associated with changes in the severity of infection.
Finally, from the experience during this study some lessons can be learned. Firstly, gathering additional data about patients diagnosed with influenza virus in the non-sentinel surveillance turned out to be extremely difficult and time consuming, because the non-sentinel surveillance is not organized to accommodate the possibility of tracing back patients or their physicians and thus collecting additional data. Consequently, the response to the questionnaire for the patients from the non-sentinel surveillance was low with only 49% for a complete dataset. Secondly, data from regular surveillance are not by definition suitable for specific analytical studies, because the objectives for data collection for surveillance might be different than for a specific analytical study. Additional study protocols will be advisable for future studies in which virological characteristics of new potentially more pathogenic influenza strains are investigated in relation to clinical and epidemiological characteristics. In the Netherlands prepandemic and pandemic study plans have been developed for such studies, that already have been used during the epidemic with influenza A(H1N1) 2009 [34Hahne S, Donker T, Meijer A, et al. Epidemiology and control of influenza A(H1N1)v in the Netherlands: the first 115 cases Eur Surveill 2009; 14(ii): 19267.]. The recent emergence of oseltamivir resistant influenza viruses in 2007-2008 as well as the recent pandemic of influenza A(H1N1) 2009 has shown that is necessary to be able to answer questions about the clinical impact of new potentially more pathogenic influenza strains or other respiratory viruses timely. Especially linked clinical and virological data from patients with severe acute respiratory infectious diseases are needed. However, specimens from hospitalized patients (i.e. non sentinel specimens) are sent in to NIC-ErasmusMC for characterization typically with minimal patient, clinical, and course of disease information and with a delay of several weeks. It has been extremely difficult to obtain robust clinical and course of disease information afterwards for these patients in this study. This phenomenon has also complicated a similar Europe-wide study in which only 5 countries, including the Netherlands, were able to participate which could obtain follow-up data relatively easy [28Ciancio BC, Meerhoff TJ, Kramarz P, et al. Oseltamivir-resistant influenza A(H1N1) viruses detected in Europe during season 2007-8 had epidemiologic and clinical characteristics similar to co-circulating susceptible A(H1N1) viruses Eur Surveill 2009; 14(ii): 19412.]. Despite pre-established plans for a pandemic we had the same experience to obtain follow-up information on hospitalized patients during the A(H1N1) 2009 pandemic. To avoid these problems in the future, the establishment of a (national) clinical network of laboratories and hospitals that surveys severe acute respiratory infectious diseases would be preferable. In such a network a close interaction between surveillance and clinical virology is needed for rapid access to combined virological and clinical data.
CONCLUSION
A large proportion influenza A(H1N1) oseltamivir resistant viruses was detected in the Netherlands in 2007-2008, whilst in our enhanced surveillance no significant effect on clinical presentation of patients was found. Hence, ORV did not confer more severe disease. This event provides justification for continuous monitoring of the antiviral drug sensitivity profile circulation of influenza viruses. In addition, more research is needed to elucidate the emergence of naturally resistant influenza viruses. The recent emergence of oseltamivir resistant influenza viruses in 2007-2008 as well as the recent pandemic of influenza A(H1N1) 2009 has shown that in the current system in the Netherlands it proved difficult to answer questions about the clinical impact of new potentially more pathogenic influenza strains or other respiratory viruses timely. Possibly, establishing a clinical network of laboratories and hospitals that surveys severe respiratory infectious diseases would fulfil this need.
CONFLICT OF INTEREST
Financial support: Dutch Ministry of Health, Welfare and Sport.
Potential conflicts of interest: Ruud van Beek, Albert Osterhaus and Guus Rimmelzwaan are part-time employed by Viroclinics Biosciences BV, a spin-off company of Erasmus MC Holding. All other authors: no conflicts.
ACKNOWLEDGEMENTS
We thank all patients and their general practitioners or hospital physicians for providing information and specimens for investigation, the Dutch virologists for sending non-sentinel virus isolates to the NIC, the Dutch Society for Clinical Virology for their help in designing the clinical study among non-sentinel patients, and Mrs. Heshusius-Valen (NIVEL) for her crucial role in data collection. We thank Wichard Tilstra, Astrid Glas, Mariam Bagheri and Shireen Jenny for technical assistance.
REFERENCES
| [1] | Simonsen L. The global impact of influenza on morbidity and mortality Vaccine 1999; 17(Suppl 1): S3-10. |
| [2] | Kramarz P, Monnet D, Nicoll A, Yilmaz C, Ciancio B. Use of oseltamivir in 12 European countries between 2002 and 2007--lack of association with the appearance of oseltamivir-resistant influenza A(H1N1) viruses Eur Surveill 2009; 14(ii): 19112. |
| [3] | Meijer A, Lackenby A, Hay A, Zambon M. Influenza antiviral susceptibility monitoring activities in relation to national antiviral stockpiles in Europe during the winter 2006/2007 season Eur Surveill 2007; 12: E3-4. |
| [4] | Jonges M, Van Der Lubben IM, Dijkstra F, Verhoef L, Koopmans M, Meijer A. Dynamics of antiviral-resistant influenza viruses in the Netherlands, 2005-2008 Antiviral Res 2009; 83: 290-7. |
| [5] | Escuret V, Frobert E, Bouscambert-Duchamp M, et al. Detection of human influenza A (H1N1) and B strains with reduced sensitivity to neuraminidase inhibitors J Clin Virol 2008; 41: 25-8. |
| [6] | Sheu TG, Deyde VM, Okomo-Adhiambo M, et al. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008 Antimicrob Agents Chemother 2008; 52: 3284-92. |
| [7] | Monto AS, McKimm-Breschkin JL, Macken C, et al. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use Antimicrob Agents Chemother 2006; 50: 2395-402. |
| [8] | Mungall BA, Xu X, Klimov A. Surveillance of influenza isolates for susceptibility to neuraminidase inhibitors during the 2000-2002 influenza seasons Virus Res 2004; 103: 195-7. |
| [9] | ECDC. Timeline for the emergence of oseltamivir resistant influenza A(H1N1) 2007-8 Available from: http://ecdc.europa.eu/en/files/pdf/Health_topics/ECDC_AVR_timeline_v1_31-10-08.pdf [cited 2008 November 21]; |
| [10] | Hauge SH, Dudman S, Borgen K, Lackenby A, Hungnes O. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007-08 Emerg Infect Dis 2009; 15: 155-62. |
| [11] | Lackenby A, Hungnes O, Dudman SG, et al. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe Eur Surveill 2008; 13(ii): 8026. |
| [12] | Aoki FY, Boivin G, Roberts N. Influenza virus susceptibility and resistance to oseltamivir Antivir Ther 2007; 12: 603-16. |
| [13] | Kiso M, Mitamura K, Sakai-Tagawa Y, et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study Lancet 2004; 364: 759-65. |
| [14] | Ward P, Small I, Smith J, Suter P, Dutkowski R. Oseltamivir (Tamiflu) and its potential for use in the event of an influenza pandemic J Antimicrob Chemother 2005; 55(Suppl 1): i5-i21. |
| [15] | Ives JA, Carr JA, Mendel DB, et al. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo Antiviral Res 2002; 55: 307-17. |
| [16] | Donker GA. Continuous morbidity registration at Dutch sentinel stations, 2007 Utrecht: NIVEL 2008. |
| [17] | Jonges M, Liu WM, van der Vries E, et al. Influenza virus inactivation for studies of antigenicity and phenotypic neuraminidase inhibitor resistance profiling J Clin Microbiol 2010; 48: 928-40. |
| [18] | Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate Anal Biochem 1979; 94: 287-96. |
| [19] | Wetherall NT, Trivedi T, Zeller J, et al. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors: report of the neuraminidase inhibitor susceptibility network J Clin Microbiol 2003; 41: 742-50. |
| [20] | De Jong JC, Rimmelzwaan GF, Bartelds AI, Wilbrink B, Fouchier RA, Osterhaus AD. [2000/01 influenza season and the vaccine composition for the season 2001/'02] Ned Tijdschr Geneeskd 2001; 145: 1945-50. |
| [21] | Dijkstra F, Donker GA, Wilbrink B, Van Gageldonk-Lafeber AB, Van Der Sande MA. Long time trends in influenza-like illness and associated determinants in the Netherlands Epidemiol Infect 2009; 137: 473-9. |
| [22] | Massart DL, Smeyers-Verbeke J, Capron X, Schlesier K. Visual presentation of data by means of box plots LC-GC Europe 2005; 18: 215-18. |
| [23] | Rimmelzwaan GF, De Jong JC, Donker GA, Meijer A, Fouchier RA, Osterhaus AD. Influenza season 2007/'08 in the Netherlands: antigenic variation, oseltamivir resistance and vaccine composition for the 2008/'09 season Ned Tijdschr Geneeskd 2008; 152: 2138-44. |
| [24] | Dharan NJ, Gubareva LV, Meyer JJ, et al. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States JAMA 2009; 301: 1034-41. |
| [25] | World Health Organization. Influenza A(H1N1) virus resistance to oseltamivir; last quarter 2007 to first quarter 2008 Available from: http://www.who.int/csr/disease/influenza/oseltamivir_summary/en/index.html [cited 2008 November 21]; |
| [26] | Besselaar TG, Naidoo D, Buys A, et al. Widespread oseltamivir resistance in influenza A viruses (H1N1), South Africa Emerg Infect Dis 2008; 14: 1809-0. |
| [27] | Meijer A, Lackenby A, Hungnes O, et al. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 Season Emerg Infect Dis 2009; 15: 552-60. |
| [28] | Ciancio BC, Meerhoff TJ, Kramarz P, et al. Oseltamivir-resistant influenza A(H1N1) viruses detected in Europe during season 2007-8 had epidemiologic and clinical characteristics similar to co-circulating susceptible A(H1N1) viruses Eur Surveill 2009; 14(ii): 19412. |
| [29] | Gooskens J, Jonges M, Claas EC, Meijer A, Van Den Broek PJ, Kroes AM. Morbidity and mortality associated with nosocomial transmission of oseltamivir-resistant influenza A(H1N1) virus JAMA 2009; 301: 1042-6. |
| [30] | Van Der Vries E, Van Den Berg B, Schutten M. Fatal oseltamivir-resistant influenza virus infection N Engl J Med 2008; 359: 1074-6. |
| [31] | Saito R, Li D, Shimomura C, et al. An off-seasonal amantadine-resistant H3N2 influenza outbreak in Japan Tohoku J Exp Med 2006; 210: 21-7. |
| [32] | Collins PJ, Haire LF, Lin YP, et al. Structural basis for oseltamivir resistance of influenza viruses Vaccine 2009; 27: 6317-23. |
| [33] | Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance Science 2010; 328: 1272-5. |
| [34] | Hahne S, Donker T, Meijer A, et al. Epidemiology and control of influenza A(H1N1)v in the Netherlands: the first 115 cases Eur Surveill 2009; 14(ii): 19267. |








