- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
Luciferase Reporter Assay System for Deciphering GPCR Pathways
Zhijie Cheng, Denise Garvin, Aileen Paguio, Pete Stecha, Keith Wood, Frank Fan*
Abstract
The G protein coupled receptors (GPCR) represent the target class for nearly half of the current therapeutic drugs and remain to be the focus of drug discovery efforts. The complexity of receptor signaling continues to evolve. It is now known that many GPCRs are coupled to multiple G-proteins, which lead to regulation of respective signaling pathways downstream. Deciphering this receptor coupling will aid our understanding of the GPCR function and ultimately developing drug candidates. Here, we report the development of four homogenous bioluminescent reporter assays using improved destabilized luciferases and various response elements: CRE, NFAT-RE, SRE, and SRF-RE. These assays allowed measurement of major GPCR pathways including cAMP production, intracellular Ca2+ mobilizations, ERK/MAPK activ-ity, and small G protein RhoA activity, respectively using the same reporter assay format. We showed that we can decipher G protein activation profiles for exogenous m3 muscarinic receptor and endogenous β2-adrenergic receptors in HEK293 cells by using these four reporter assays. Furthermore, we demonstrated that these assays can be readily used for potency rankings of agonists and antagonists, and for high throughput screening.
Article Information
Identifiers and Pagination:
Year: 2010Volume: 4
First Page: 84
Last Page: 91
Publisher Id: CCGTM-4-84
DOI: 10.2174/1875397301004010084
Article History:
Received Date: 21/8/2010Revision Received Date: 25/10/2010
Acceptance Date: 25/10/2010
Electronic publication date: 21/12/2010
Collection year: 2010
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.5/), which permits unrestrictive use, distribution, and reproduction in any medium, provided the original work is properly cited.
* Address correspondence to this author at the Department of Research, Promega Corporation, Madison, WI53711, USA; Tel: 608-277-2531; E-mail: frank.fan@promega.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 21-8-2010 |
Original Manuscript | Luciferase Reporter Assay System for Deciphering GPCR Pathways | |
INTRODUCTION
G protein coupled receptors (GPCRs, or 7TM receptors), constitute the largest superfamily of cell surface receptors and play key roles in regulation of wide variety of physiological processes, including cognition, metabolism, inflammation, immunity and cell proliferation and development [1Lefkowitz RJ. Seven transmembrane receptors: something old, something new Acta Physiol (Oxf) 2007; 190: 9-19.]. As a result, GPCRs have been popular drug targets, and nearly half of approved drugs on the market directly target GPCRs. Despite the large number of GPCRs that are available as potential drug targets, currently approved drugs only address a small portion of receptors while hundreds are under development. Thus, this target class remains of strong interest for drug discovery.
GPCRs transmit extracellular signals across the plasma membrane via intracellular coupling with heterotrimeric G proteins. Heterotrimeric G proteins are classified into four subfamilies based on their Gα subunit, Gs, Gi, Gq and G12. Upon activation of GPCRs, G protein α subunit dissociates from βã dimeric subunit, which in turn initiates a cascade of downstream second messenger pathways and eventually induces gene transcription by various response elements including cAMP response element (CRE), nuclear factor of activated T-cells response element (NFAT-RE), serum response element (SRE) and serum response factor response element (SRF-RE, a mutant form of SRE) (Fig. 1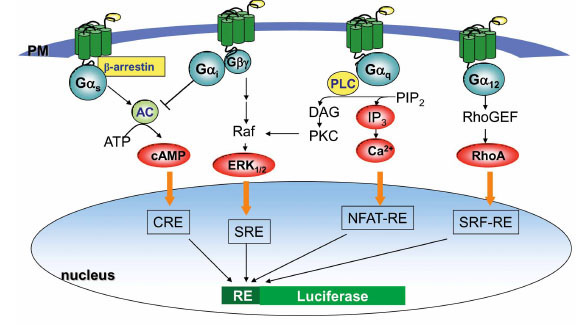 ). Over the past few decades there have been many substantial discoveries regarding GPCRs structure and function including β-arrestin-dependent signaling, allosteric modulation, functional selectivity and receptor dimerization that have led to the current understanding of the complexity of receptor signaling [2Shenoy SK, Drake MT, Nelson CD, et al. beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor J Biol Chem 2006; 281: 1261-73.-4Chan WY, McKinzie DL, Bose S, et al. Allosteric modulation of the muscarinic M4 receptor as an approach to treating schizophrenia Proc Natl Acad Sci U S A 2008; 105: 10978-83.]. Some GPCRs only couple to one type of G protein (e.g., Gs), but many GPCRs are known to couple to a broad range of G-protein families, such as Gi/G12, Gq/G12, or Gi/Gq/G12 in a cell type-dependent or compound-specific manner [5Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions Physiol Rev 2005; 85: 1159-204.]. Current methods employed in GPCR screening programs measure G protein signaling by determining change in second messengers such as cAMP, inositol trisphosphate (IP3), and intracellular Ca2+ mobilizations, which often demands setting up different assay platforms and requires specialized instrumentation for each pathway which could be costly [6Siehler S. Cell-based assays in GPCR drug discovery Biotechnol J 2008; 3: 471-83.]. Plus, HTS-compatible methods for G12-RhoA detection is lacking. Thus, despite of the demands of screening assays for multiple functional readouts, the practical execution for drug screening has been slow.
). Over the past few decades there have been many substantial discoveries regarding GPCRs structure and function including β-arrestin-dependent signaling, allosteric modulation, functional selectivity and receptor dimerization that have led to the current understanding of the complexity of receptor signaling [2Shenoy SK, Drake MT, Nelson CD, et al. beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor J Biol Chem 2006; 281: 1261-73.-4Chan WY, McKinzie DL, Bose S, et al. Allosteric modulation of the muscarinic M4 receptor as an approach to treating schizophrenia Proc Natl Acad Sci U S A 2008; 105: 10978-83.]. Some GPCRs only couple to one type of G protein (e.g., Gs), but many GPCRs are known to couple to a broad range of G-protein families, such as Gi/G12, Gq/G12, or Gi/Gq/G12 in a cell type-dependent or compound-specific manner [5Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions Physiol Rev 2005; 85: 1159-204.]. Current methods employed in GPCR screening programs measure G protein signaling by determining change in second messengers such as cAMP, inositol trisphosphate (IP3), and intracellular Ca2+ mobilizations, which often demands setting up different assay platforms and requires specialized instrumentation for each pathway which could be costly [6Siehler S. Cell-based assays in GPCR drug discovery Biotechnol J 2008; 3: 471-83.]. Plus, HTS-compatible methods for G12-RhoA detection is lacking. Thus, despite of the demands of screening assays for multiple functional readouts, the practical execution for drug screening has been slow.
Reporter gene system is a technology where a reporter gene is synthesized in response to activation of a specific signaling cascade of interests, followed by monitoring of the reporter protein expression by its enzymatic activities linked with a variety of colormetric, fluorescent or luminescent read-outs. Due to its inherent sensitivity, large signal dynamics and simplicity to set up, reporter assay platform has been widely used as high throughput homogenous assay for screening GPCR targets linked to cAMP or Ca2+ signaling [7Bronstein I, Fortin J, Stanley PE, Stewart GS, Kricka LJ. Chemiluminescent and bioluminescent reporter gene assays Anal Biochem 1994; 219: 169-81.]. Despite the success, some concerns were raised. These include the requirement of a long induction time, false positives and the fact that signal event is distal from receptor activation. These concerns have been addressed in a number of approaches such as the use of destabilized reporters and co-expression of a constitutively expressed internal control [7Bronstein I, Fortin J, Stanley PE, Stewart GS, Kricka LJ. Chemiluminescent and bioluminescent reporter gene assays Anal Biochem 1994; 219: 169-81., 8Fan F, Wood KV. Bioluminescent assays for high-throughput screening Assay Drug Dev Technol 2007; 5: 127-36.]. The high number of false positives due to the fact that the signaling event measured is so distal from receptor activation could be minimized by coexpression of a constitutively expressed internal control reporter [8Fan F, Wood KV. Bioluminescent assays for high-throughput screening Assay Drug Dev Technol 2007; 5: 127-36.]. Here, we report the development of a series of homogenous bioluminescent reporter assays using reporter vectors with CRE-, NFAT-RE-, SRE-, SRF-RE- built in upstream of destabilized luciferase gene to measure four major GPCR signaling events: change in cAMP level, Ca2+ mobilization, ERK/MAPK activity and small G protein RhoA activation. By using various reporter vectors together, we can measure receptor/G protein coupling profiles for individual receptors, and further compare the potency rankings of various compounds for the same receptor to activate multiple signal pathways. Thus, luciferase reporter assay provides an efficient approach to decipher major GPCR pathways in one simple assay format, which can be used to develop robust HTS assays for drug discovery.
MATERIALS AND METHODOLOGY
Materials
pGL4.26[luc2/Hygro], pGL4.27[luc2P/Hygro], pGL4.28 [luc2CP/Hygro], pGL4.29[CRE/minP/luc2P], pGL4.30 [NFAT-RE/ minP/luc2P], and pF9A CMV hRluc-neo Flexi vectors, and PMA were from Promega (Madison, WI). All other reagents were from Sigma (St. Louis, MO).
Plasmid Construction
The DNA sequences of five copies of the SRE or SRF-RE were synthesized by Integrated DNA Technologies, Inc. (Coralville, Iowa). SRE- and SRF-RE- reporter were constructed by insertion of SRE or SRF-RE into pGL4.26, pGL4.27, or pGL4.28 using Hind III and Bgl II restriction enzyme sites. The resulting constructs from pGL4.27 were named as pGL4.33 [SRE/luc2P] and pGL4.34[SRF-RE/ luc2P], respectively.
The human dopamine D1 receptor (accession number NM_000794) was obtained from American Type Culture Collection (Manassas, VA). The m3 muscarinic receptor (NM_000740.2) was acquired from Origene (Rockville, MD). The coding sequences of both receptors were subcloned into pF9A CMV hRluc-neo Flexi Vector to make receptor-Renilla fusion proteins. The constructs for m4 muscarinic receptor were from UMR cDNA Resource Center (Rolla, MO). All sequences were verified by restriction digest and sequencing.
Cell Culture, Transfection and Generation of Stable HEK293 Cell Lines
HEK293 cells were cultured in DMEM (Life Technologies) supplemented with 10% fetal bovine serum at 37oC with a humidified atmosphere at 5% CO2. Transient tranfections were done using TransIT®-LT1 Transfection Reagent following manufacturer’s protocol (Mirus Bio). Cells were directly plated and transfected in 96-well plate, or plated and transfected in T75 flask, then plated into 384-well plate 4-6 hours after transfection. The cells were changed to serum-starving medium 4-6 hours after transfection and assayed 20 hours later for SRE- or SRF-RE- reporter assay, or assayed 24 hours after transfection for CRE- or NFAT-RE- reporter assay.
Stable HEK293 cell lines expressing reporter vectors (CRE-, NFAT-RE, SRE-, SRF-RE-) were generated by transient transfection followed by hygromycin selection and limiting dilution cloning. Double stable cell lines co-expressing reporter vector and receptor (D1 dopamine receptor and m3 muscarinic receptor) were generated by transfection of receptor construct into single stable line expressing corresponding reporter vector. Cells were plated in complete medium for CRE- or NFAT-RE- assay, or plated in DMEM plus 2% charcoal/dextran treated FBS (Thermo Scientific) for SRE- or SRF-RE- assay 24 hours before assay.
Luciferase Reporter Assays
Firefly and renilla luciferase activities, as indicated by relative luminescence units (RLU) were determined using One-Glo or Dual-Glo luciferase assay kits (Promega) according to the manufacturer's instructions. For agonist, fold of induction = firefly RLUinduced / firefly RLUuninduced. For antagonist, % of control=100 × firefly RLU(agonist+antagonist) / firefly RLUagonist alone, all normalized to renilla RLU. Both EC50 and IC50 values were generated using GraphPad Prism software. Z’ values were determined as Z’=1- [(3×SDinudced + 3×SDuninduced) / (averageinduced – averageuninduced)">(3×SDinudced + 3×SDuninduced) / (averageinduced – averageuninduced)].
RESULTS AND DISCUSSION
Improved Assay Performance by Using Destabilized Luciferase Reporter
Previously, we have shown that the luciferase reporter vectors (pGL4 series) containing minimal promoter and destabilized luciferase gene improve the responsiveness of cAMP response element to Gs coupled receptor [8Fan F, Wood KV. Bioluminescent assays for high-throughput screening Assay Drug Dev Technol 2007; 5: 127-36.]. To evaluate if this is also applicable for other response elements which are specific for different signal pathways, we constructed SRE- and SRF-RE- reporter in various pGL4 vectors containing luciferase gene with or without protein degradation sequence hPEST (Pro-Glu-Ser-Thr) and CL1 [9Li X, Zhao X, Fang Y, et al. Generation of destabilized green fluorescent protein as a transcription reporter J Biol Chem 1998; 273: 34970-5., 10Gilon T, Chomsky O, Kulka RG. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae EMBO J 1998; 17: 2759-66.] as shown in Fig. ( 2A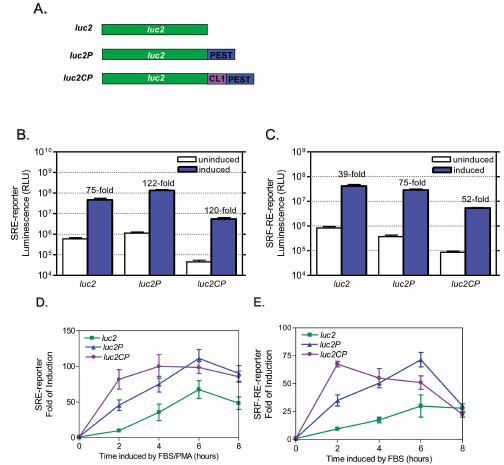 ). SRE is known to respond to ternary complex factor (TCF)-dependent ERK/MAPK pathway, while SRF-RE, a mutant form of SRE lacking TCF binding domain, is newly designed to respond to SRF-dependent and TCF-independent pathway such as RhoA activation [11Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF Cell 1995; 81: 1159-70.]. GPCRs, particularly those coupled with Gi and Gq activate ERK/MAPK pathway and induce transcriptional activation of SRE, while GPCRs coupled with G12 family are known to activate Rho guanine nucleotide exchange factors (RhoGEFs) which leads to activation of RhoA and transcriptional activation of SRF-RE [12Hill CS, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors EMBO J 1995; 14: 5037-47.]. This is particularly important with increasing interests in HTS drug screening targeting G12/RhoA pathway while there are no HTS-compatible methods available for G12/RhoA so far.
). SRE is known to respond to ternary complex factor (TCF)-dependent ERK/MAPK pathway, while SRF-RE, a mutant form of SRE lacking TCF binding domain, is newly designed to respond to SRF-dependent and TCF-independent pathway such as RhoA activation [11Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF Cell 1995; 81: 1159-70.]. GPCRs, particularly those coupled with Gi and Gq activate ERK/MAPK pathway and induce transcriptional activation of SRE, while GPCRs coupled with G12 family are known to activate Rho guanine nucleotide exchange factors (RhoGEFs) which leads to activation of RhoA and transcriptional activation of SRF-RE [12Hill CS, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors EMBO J 1995; 14: 5037-47.]. This is particularly important with increasing interests in HTS drug screening targeting G12/RhoA pathway while there are no HTS-compatible methods available for G12/RhoA so far.
SRE-mediated gene transcription is known to be rapidly induced by serum, phorbol esters and growth factors in TCF and SRF-dependent manner, while SRF alone can mediate transcription activation of SRF-RE by serum mostly through the binding to cell surface receptors of the mitogenic agents such as LPA in serum [11Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF Cell 1995; 81: 1159-70., 12Hill CS, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors EMBO J 1995; 14: 5037-47.]. Thus, to establish our assay we first used serum plus PMA to stimulate SRE reporter, and used serum alone to simulate SRF-RE reporter. When various SRE- or SRF-RE reporters were transiently transfected into HEK293 cells, the luciferase reporters containing protein degradation sequence hPEST (luc2P), or both hPEST and CL1 (luc2CP) showed 5-10 times lower assay background due to decreased basal expression level of reporter proteins (Fig. 2B , 2C
, 2C ). The cells were further induced with FBS plus PMA (for SRE-) or FBS alone (for SRF-RE-) for different time points up to 24 hours (data not shown for 8+ hour time point). As shown in Fig. (2D
). The cells were further induced with FBS plus PMA (for SRE-) or FBS alone (for SRF-RE-) for different time points up to 24 hours (data not shown for 8+ hour time point). As shown in Fig. (2D and 2E
and 2E ), the destabilized SRE- and SRF-RE- reporters reached the maximum of response in a shorter time (2-6 hours) than traditional luciferase gene (8+ hours).
), the destabilized SRE- and SRF-RE- reporters reached the maximum of response in a shorter time (2-6 hours) than traditional luciferase gene (8+ hours).
The long incubation time required by traditional reporter assays has limited the application of reporter technology in HTS drug screening due to the concerns of cytotoxicity by the compounds. Therefore, deployment of the destabilized reporter gene will remove this hurdle with improved response dynamics and reduced assay time (2-6 hours) which in turn could potentially minimize secondary effects (such as toxicity effect) arising from the prolonged incubation of cells with chemical compounds. Here we have shown that Luc2P versions of SRE and SRF-RE reached the peak response in six hours (Fig. 2D , 2E
, 2E ) with minimal sacrifice of basal reporter expression (Fig. 2B
) with minimal sacrifice of basal reporter expression (Fig. 2B , 2C
, 2C ). Similar results were observed when we further measured several GPCR responses by comparing these SRE- or SRF-RE reporters with or without protein degradation sequences (data not shown), confirming the potentials of using destabilized luciferase reporters in GPCR drug screening. Thus, in consideration of both assay dynamics and signal output, we chose the reporter vectors with protein degradation sequence hPEST incorporated with luciferase, SRE-luc2P and SRF-RE-luc2P and six hour induction time for most of the following evaluation, unless otherwise indicated.
). Similar results were observed when we further measured several GPCR responses by comparing these SRE- or SRF-RE reporters with or without protein degradation sequences (data not shown), confirming the potentials of using destabilized luciferase reporters in GPCR drug screening. Thus, in consideration of both assay dynamics and signal output, we chose the reporter vectors with protein degradation sequence hPEST incorporated with luciferase, SRE-luc2P and SRF-RE-luc2P and six hour induction time for most of the following evaluation, unless otherwise indicated.
Amenable for High Throughput Screening
To determine if luciferase reporter assay is amenable for high-throughput screening for GPCR modulators linked with all major G protein signaling, we chose four individual receptors including D1 dopamine receptor, m4 muscarinic receptor, m3 muscarinic receptor and endogenous EDG receptor, which are known to couple with Gs, Gi, Gq and G12 subfamily respectively, and co-expressed them with the corresponding reporters. In the cases of Gs, Gq and G12 signaling, luc2P version of reporters (CRE-luc2P, NFAT-RE-luc2P, SRF-RE-luc2P) were tested and shown to give maximal assay performance, while luc2CP of SRE reporter was used for detection of Giβγ-linked MAP kinase activation in order to achieve optimized assay dynamics (Fig. 3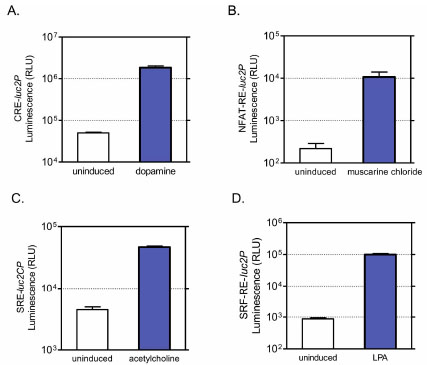 , Table 1).
, Table 1).
Cells expressing luciferase reporter and receptors were induced in a 384-well plate with various agonists for two to eight hours depending on the requirement of the response elements, RLUs were measured and Z’-factor values were then determined. Z’-factor is one of the major statistical parameters commonly used to evaluate assay performance in high throughput screening. According to the formula, the closer the Z’-factor is to 1, the better the assay quality is. In general, Z’ values greater than 0.5 are acceptable for HTS [13Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays J Biomol Screen 1999; 4: 67-73.]. As shown in Table T11, all four reporter assays showed Z’-factor values significantly higher than 0.5 in 384-well format with large response dynamic (fold of induction) range from 10 to 300 fold. Thus, luciferase reporter assays are robust, and can be used for high throughput screening of modulators for all major G protein pathways.
Deciphering GPCR Coupled Signaling Pathways
We next evaluate if luciferase reporter assay can be used to profile receptor/G protein coupling in HEK293 cells. It is known that the m3 muscarinic receptor, when exogenously expressed, couples with both Gq and G12 pathways, while endogenously expressed β2-adrenergic receptors mostly activate Gs protein. When we overexpressed m3 muscarinic receptor in cells in four individual reporter assays, receptor activation by agonist carbachol induced dose-dependent expression of luciferase with distinguishable potencies (Fig. 4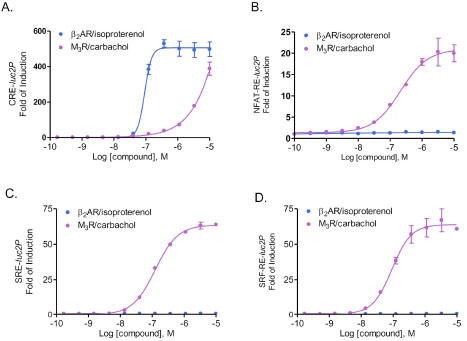 ). As indicated in Table 2, NFAT-RE-, SRE- and SRF-RE assays showed similar EC50 values by carbachol stimulation, suggesting that activation of m3 muscarinic receptors leads to increase of intracellular calcium concentration, MAP kinase and RhoA activation with similar efficacies. In contrast, EC50 values shown by CRE- reporter assay was almost three logs higher than those by other reporter assays tested, indicating that m3 muscarinic receptors activate cAMP production in a much less efficient manner. In contrast, activation of β2-adrenergic receptors by isoproterenol showed dose-dependent response in CRE-reporter assay, but not in NFAT-RE-, SRE- and SRF-RE- reporter assays, confirming that the major downstream signaling for β2-adrenergic receptors is cAMP pathway via Gs protein. Thus, luciferase reporter technology is able to study G protein activation profiles by the same receptor in one single assay format.
). As indicated in Table 2, NFAT-RE-, SRE- and SRF-RE assays showed similar EC50 values by carbachol stimulation, suggesting that activation of m3 muscarinic receptors leads to increase of intracellular calcium concentration, MAP kinase and RhoA activation with similar efficacies. In contrast, EC50 values shown by CRE- reporter assay was almost three logs higher than those by other reporter assays tested, indicating that m3 muscarinic receptors activate cAMP production in a much less efficient manner. In contrast, activation of β2-adrenergic receptors by isoproterenol showed dose-dependent response in CRE-reporter assay, but not in NFAT-RE-, SRE- and SRF-RE- reporter assays, confirming that the major downstream signaling for β2-adrenergic receptors is cAMP pathway via Gs protein. Thus, luciferase reporter technology is able to study G protein activation profiles by the same receptor in one single assay format.
Many GPCRs activate a variety of signaling pathways via distinct G proteins in cell type dependent manner, which is crucial for their precise regulation of tissue-specific physiological functions [14Castaño JP, Martínez-Fuentes AJ, Gutiérrez-Pascual E, et al. Intracellular signaling pathways activated by kisspeptins through GPR54: do multiple signals underlie function diversity? Peptides 2009; 30: 10-5.]. Unfortunately many of the current available screening methods have limitations and are unable to measure multiple GPCR signaling events in one assay format. Also, not all GPCR signaling such as RhoA activity can be measured in HTS format. Thus, despite significant interest in profiling for cell type dependent receptor/G protein coupling, it has been progressed rather slowly in current drug discovery practice, partly due to the burden setting up multiple assay platforms and lack of existing detection tools. The bioluminescent reporter assay using destabilized luciferase is fast, highly sensitive and easy to set up. More importantly, as we have shown here in Fig. (4 ), bioluminescent reporter assays can measure all major GPCR signaling via Gs, Gi, Gq and G12 pathways in one single assay format with great assay dynamics and Z’ values, thereby they could be potentially used to monitor cell type-dependent signaling via G protein coupled receptors.
), bioluminescent reporter assays can measure all major GPCR signaling via Gs, Gi, Gq and G12 pathways in one single assay format with great assay dynamics and Z’ values, thereby they could be potentially used to monitor cell type-dependent signaling via G protein coupled receptors.
Potency Ranking of GPCR Modulators for Different G Protein Pathways
Since NFAT-RE- or SRE-luc2P reporters respond nicely to activation of m3 muscarinic receptor (Fig.4 ), we next compared the potency ranking of a series of well-known compounds for m3 muscarinic receptor using both reporter assays. As expected, co-transfection of either reporter vector with m3 muscarinic receptor showed a large dynamic range of assay response upon agonist stimulation and both reporter assays showed identical orders of potency ranking for the agonists and antagonists we tested (Fig. 5
), we next compared the potency ranking of a series of well-known compounds for m3 muscarinic receptor using both reporter assays. As expected, co-transfection of either reporter vector with m3 muscarinic receptor showed a large dynamic range of assay response upon agonist stimulation and both reporter assays showed identical orders of potency ranking for the agonists and antagonists we tested (Fig. 5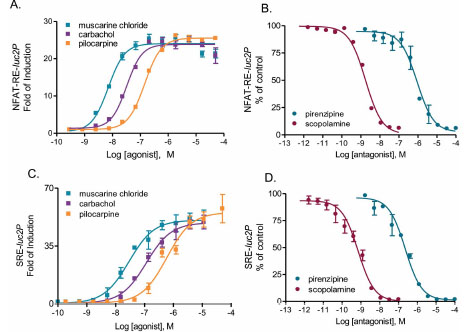 ). The order of potency rankings for agonists were: muscarine chloride>carbachol>pilocarpine; The IC50s for antagonists were ranked as: scopolamine>pirenzipine, in the presence of muscarine chloride. Both potency rankings are consistent with those reported in the literature (Table 3). The potencies of same compound/receptor pairs attained from two reporter assays are all within a half-log range, suggesting none of the compounds showed functional selectivity (Fig. 5
). The order of potency rankings for agonists were: muscarine chloride>carbachol>pilocarpine; The IC50s for antagonists were ranked as: scopolamine>pirenzipine, in the presence of muscarine chloride. Both potency rankings are consistent with those reported in the literature (Table 3). The potencies of same compound/receptor pairs attained from two reporter assays are all within a half-log range, suggesting none of the compounds showed functional selectivity (Fig. 5 ).
).
More evidence indicates that same receptor could activate various signaling pathways with distinct potencies depending on the modulators which could bind to the receptor orthosterically or allosterically. This new concept offers great potential for developing pathway-specific drugs that increase efficacy and reduce side effects, but at the same time add another dimension in assay complexity to drug discovery. In the case of m3 muscarinic receptor, activation of m3 muscarinic receptor increases intracellular Ca2+ mobilization and induces ERK/MAPK pathway at the same time, however, it is also reported that MAPK activity via m3 muscarinic receptor is regulated in a calcium-dependent or independent manner in a variety cell types, suggesting the involvement of distinct signal mechanisms [15Slack BE. The m3 muscarinic acetylcholine receptor is coupled to mitogen-activated protein kinase via protein kinase C and epidermal growth factor receptor kinase Biochem J 2000; 348: 381-7.]. Therefore, by comparing the potencies of same compound/receptor pairs using multiple reporter vectors which measure different G protein downstream signaling, this approach has great potential in drug screening for GPCR modulators with functional selectivity in one simple assay format.
CONCLUSION
In this study, we developed four homogenous bioluminescent reporter assays using destabilized luciferase. The improved reporter assays showed larger assay dynamics in shorter assay time than using regular luciferase. Using a model system of exogenous or endogenous receptors in HEK293 cells, we demonstrated that all four major G protein subfamilies and downstream pathways can be studied in one luciferase reporter assay format. Furthermore, these assays can be readily used for potency rankings of GPCR modulators and for high throughput screening. Together, our data suggest that these improved luciferase reporter assays are well suited for GPCR research and drug screening, with potential application in establishing receptor/G protein profiles for any particular receptor and drug screening for pathway-specific GPCR modulators.
ACKNOWLEDGEMENT
We would like to thank Kevin Kopish for valuable discussion.